Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Immunocytochemistry [1]
- Immunohistochemistry [1]
- Other assay [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-64020 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- PES1 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant protein fragment
- Description
- Immunogen sequence: QWVFDSVNAR LLLPVAEYFS GVQLPPHLSP FVTEKEGDYV PPEKLKLLAL QRGEDPGNLN ESEEE Highest antigen sequence identity to the following orthologs: Mouse - 88%, Rat - 89%.
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 0.2 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references Identification of the toxic 6mer seed consensus for human cancer cells.
Patel M, Bartom ET, Paudel B, Kocherginsky M, O'Shea KL, Murmann AE, Peter ME
Scientific reports 2022 Mar 24;12(1):5130
Scientific reports 2022 Mar 24;12(1):5130
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
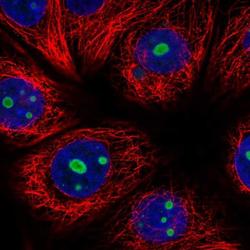
- Experimental details
- Immunofluorescent staining of PES1 in human cell line MCF7 shows positivity in nucleoli. Samples were probed using a PES1 Polyclonal Antibody (Product # PA5-64020).
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
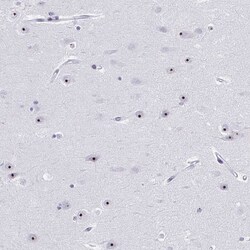
- Experimental details
- Immunohistochemical analysis of PES1 in human hippocampus using PES1 Polyclonal Antibody (Product # PA5-64020) shows strong nucleolar positivity in neuronal cells.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
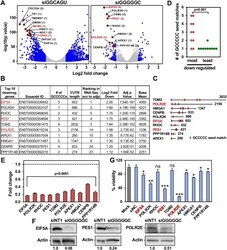
- Experimental details
- Figure 4 siGGGGGC kills cells by targeting GCCCCC seed matches located predominantly in the 3' UTR of targeted mRNAs. ( A ) Volcano plot of deregulated genes in HeyA8 cells transfected with 10 nM siGGGGGC vs. siNT1 (after 24 h) or with 10 nM siGGCAGU vs. siNT1 (after 48 h). Red dots indicate genes that were most highly expressed (>1000 norm reads) and down regulated. The number of GCCCCC seed matches is given in brackets. Grey dots indicate all differentially expressed genes (with a log2 fold change < 3 and > - 3), blue dots indicate genes with log2 fold change > 1 and FDRAdjpValue < 0.05. ( B ) Table showing the 10 downregulated genes labeled in A ranked (from top to bottom) according to Log2 fold change. ( C ) Schematic of the 3' UTRs of the 10 genes listed in B with the location of the GCCCCC seed matches. ( D ) The number of predicted seed matches in the ten most and least downregulated and highly expressed genes part of the 5957 genes in the human genome that contain at least one such seed match in their 3' UTR. p-value was calculated by Student's t test. ( E ) Real-time PCR validation of downregulation of the ten genes in ( B ) derived from RNA seq analysis (24 h time point). Gene expression levels in HeyA8 cells transfected with siGGGGGC were normalized to GAPDH and to cells transfected with siNT1 (stippled line). Each bar represents +- SD of three replicates. Student's t test was used to calculate p-values for each gene. ( F ) Western blot analysis for EIF5A, POLR2E,
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry