Antibody data
- Antibody Data
- Antigen structure
- References [4]
- Comments [0]
- Validations
- Western blot [1]
- Immunohistochemistry [1]
- Chromatin Immunoprecipitation [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA003908 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA003908, RRID:AB_1080063
- Product name
- Anti-SOX6
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human SOX6, Gene description: SRY (sex determining region Y)-box 6, Validated applications: WB, IHC, ChIP, Uniprot ID: P35712, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references Systematic multi-omics cell line profiling uncovers principles of Ewing sarcoma fusion oncogene-mediated gene regulation.
Oncogenic hijacking of a developmental transcription factor evokes vulnerability toward oxidative stress in Ewing sarcoma.
Systematic validation of antibody binding and protein subcellular localization using siRNA and confocal microscopy
Myh7b/miR-499 gene expression is transcriptionally regulated by MRFs and Eos
Orth MF, Surdez D, Faehling T, Ehlers AC, Marchetto A, Grossetête S, Volckmann R, Zwijnenburg DA, Gerke JS, Zaidi S, Alonso J, Sastre A, Baulande S, Sill M, Cidre-Aranaz F, Ohmura S, Kirchner T, Hauck SM, Reischl E, Gymrek M, Pfister SM, Strauch K, Koster J, Delattre O, Grünewald TGP
Cell reports 2022 Dec 6;41(10):111761
Cell reports 2022 Dec 6;41(10):111761
Oncogenic hijacking of a developmental transcription factor evokes vulnerability toward oxidative stress in Ewing sarcoma.
Marchetto A, Ohmura S, Orth MF, Knott MML, Colombo MV, Arrigoni C, Bardinet V, Saucier D, Wehweck FS, Li J, Stein S, Gerke JS, Baldauf MC, Musa J, Dallmayer M, Romero-Pérez L, Hölting TLB, Amatruda JF, Cossarizza A, Henssen AG, Kirchner T, Moretti M, Cidre-Aranaz F, Sannino G, Grünewald TGP
Nature communications 2020 May 15;11(1):2423
Nature communications 2020 May 15;11(1):2423
Systematic validation of antibody binding and protein subcellular localization using siRNA and confocal microscopy
Stadler C, Hjelmare M, Neumann B, Jonasson K, Pepperkok R, Uhlén M, Lundberg E
Journal of Proteomics 2012;75(7):2236-2251
Journal of Proteomics 2012;75(7):2236-2251
Myh7b/miR-499 gene expression is transcriptionally regulated by MRFs and Eos
Yeung F, Chung E, Guess M, Bell M, Leinwand L
Nucleic Acids Research 2012;40(15):7303-7318
Nucleic Acids Research 2012;40(15):7303-7318
No comments: Submit comment
Enhanced validation
- Submitted by
- Atlas Antibodies (provider)
- Enhanced method
- Recombinant expression validation
- Main image
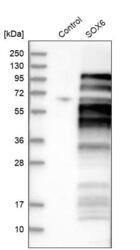
- Experimental details
- Western blot analysis in control (vector only transfected HEK293T lysate) and SOX6 over-expression lysate (Co-expressed with a C-terminal myc-DDK tag (~3.1 kDa) in mammalian HEK293T cells, LY403243).
- Sample type
- Human
- Protocol
- Protocol
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Enhanced method
- Orthogonal validation
- Main image
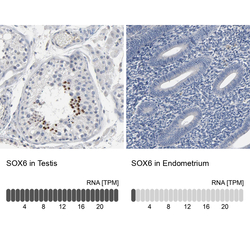
- Experimental details
- Immunohistochemistry analysis in human testis and endometrium tissues using HPA003908 antibody. Corresponding SOX6 RNA-seq data are presented for the same tissues.
- Sample type
- Human
- Protocol
- Protocol
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
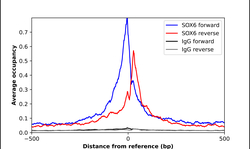
- Experimental details
- ChIP-Exo-Seq composite graph for Anti-SOX6 (HPA003908, Lot A37311) tested in K562 cells. Strand-specific reads (blue: forward, red: reverse) and IgG controls (black: forward, grey: reverse) are plotted against the distance from a composite set of reference binding sites. The antibody exhibits robust target enrichment compared to a non-specific IgG control and precisely reveals its structural organization around the binding site. Data generated by Prof. B. F. Pugh´s Lab at Cornell University.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunohistochemistry
Immunohistochemistry