Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Immunohistochemistry [3]
- Other assay [2]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-32132 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- OXSM Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant full-length protein
- Description
- Recommended positive controls: HeLa. Predicted reactivity: Mouse (88%), Rat (88%), Bovine (84%). Store product as a concentrated solution. Centrifuge briefly prior to opening the vial.
- Reactivity
- Human, Mouse, Rat
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 1 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references Mitochondrial fatty acid synthesis coordinates oxidative metabolism in mammalian mitochondria.
Nowinski SM, Solmonson A, Rusin SF, Maschek JA, Bensard CL, Fogarty S, Jeong MY, Lettlova S, Berg JA, Morgan JT, Ouyang Y, Naylor BC, Paulo JA, Funai K, Cox JE, Gygi SP, Winge DR, DeBerardinis RJ, Rutter J
eLife 2020 Aug 17;9
eLife 2020 Aug 17;9
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
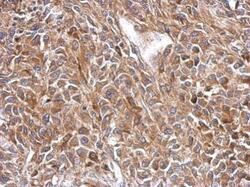
- Experimental details
- Immunohistochemical analysis of paraffin-embedded RT2 xenograft, using OXSM (Product # PA5-32132) antibody at 1:500 dilution. Antigen Retrieval: EDTA based buffer, pH 8.0, 15 min.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
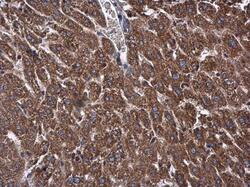
- Experimental details
- OXSM Polyclonal Antibody detects OXSM protein at mitochondria in mouse liver by immunohistochemical analysis. Sample: Paraffin-embedded mouse liver. OXSM Polyclonal Antibody (Product # PA5-32132) diluted at 1:500. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
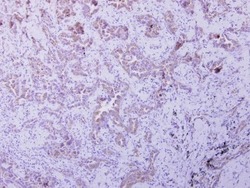
- Experimental details
- OXSM Polyclonal Antibody detects OXSM protein at cytoplasm on human lung carcinoma by immunohistochemical analysis. Sample: Paraffin-embedded lung carcinoma. OXSM Polyclonal Antibody (Product # PA5-32132) dilution: 1:250. Antigen Retrieval: EDTA based buffer, pH 8.0, 15 min.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
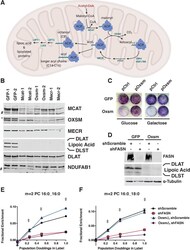
- Experimental details
- Figure 1. MtFAS is an essential pathway in mammalian skeletal myoblasts but does not contribute to synthesis of major cellular lipids. ( A ) Schematic of the mitochondrial fatty acid synthesis pathway and downstream lipoic acid synthesis. ( B ) Crude isolated mitochondrial fractions from duplicate single cell clones of Mcat , Oxsm , and Mecr mutants, compared with GFP control clonal cell lines, were separated via SDS-PAGE and immunoblotted for the indicated targets. *=Lipoic acid band (reprobe of earlier blot). #=non specific bands C . GFP control and Oxsm mutant cells were infected with retroviral control plasmid (pCtrl) or a plasmid expressing Oxsm off the CMV promoter (pOxsm), plated at equal densities in normal growth medium with either 4.5 g/L glucose or 10 mM galactose and grown for 3 or 4 days, respectively, then stained with crystal violet. ( D ) Whole cell lysates from stable cell lines generated by infecting Oxsm mutant cells (OxsmDelta) or GFP controls with shRNA constructs targeting FASN (shFASN) or scramble control (shScramble) were separated by SDS-PAGE and immunoblotted for FASN, lipoylated proteins, or tubulin. ( E-F ) Stable cell lines created in ( D ) were incubated with U 13 C-glucose for the indicated number of doublings, harvested, lipids extracted, and analyzed via LC-MS. Shown are quantitation of m+2 isotopologues for two representative phospholipid species, PC 16:0_16:0 and PC 16:0_18:0. +=p < 0.001, ++=p < 0.0001, error bars are SEM. Figure 1--figure
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
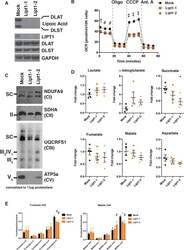
- Figure 5. MtFAS mutant phenotypes are not recapitulated by loss of lipoic acid alone. ( A ) Immunoblot for the indicated proteins in whole cell lysates from Lipt1 mutant and control cell lines. ( B ) In triplicate experiments, cells of each of the indicated clones were seeded in eight wells of a 96-well seahorse plate and allowed to adhere overnight, then equilibrated and treated with the indicated drugs following standard mitochondrial stress test protocols from the manufacturer to determine Oxygen Consumption Rate (OCR). #=p < 0.01, +=p < 0.001, ++=p < 0.0001 all comparisons are to GFP control, error bars are SEM. ( C ) Mitochondrial lysates generated from the indicated cell lines by differential centrifugation were normalized for total protein by BCA assay, incubated with 1% digitonin, then separated by BN-PAGE and immunoblotted with the indicated antibodies. ( D ) Four biological replicates from mtFAS mutant cell lines or GFP control were grown under standard proliferative conditions and harvested for steady-state metabolomics analysis by LC-MS. Shown are relative pool sizes for the indicated metabolites. *=p < 0.05, error bars are SD. ( E ) Four biological samples of the indicated genotype were labeled for 24 hr with U 13 Cglutamine, harvested, and analyzed via GC-MS for the indicated metabolites and their isotopologues. *=p < 0.05, #=p < 0.01, +=p < 0.001, error bars are SD. Figure 5--figure supplement 1. Expression of mtFAS proteins is unchanged in Lipt1 mutant cell li
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunohistochemistry
Immunohistochemistry