Antibody data
- Antibody Data
- Antigen structure
- References [32]
- Comments [0]
- Validations
- Other assay [19]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 37-1100 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- UBC13 Monoclonal Antibody (4E 11)
- Antibody type
- Monoclonal
- Antigen
- Recombinant full-length protein
- Reactivity
- Human, Mouse, Rat
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- 4E 11
- Vial size
- 100 μg
- Concentration
- 0.5 mg/mL
- Storage
- -20°C
Submitted references Ubc13 Promotes K63-Linked Polyubiquitination of NLRP3 to Activate Inflammasome.
Abraxas suppresses DNA end resection and limits break-induced replication by controlling SLX4/MUS81 chromatin loading in response to TOP1 inhibitor-induced DNA damage.
The lncRNA 'UCA1' modulates the response to chemotherapy of ovarian cancer through direct binding to miR-27a-5p and control of UBE2N levels.
Immune regulator LGP2 targets Ubc13/UBE2N to mediate widespread interference with K63 polyubiquitination and NF-κB activation.
Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination.
Advanced Cataloging of Lysine-63 Polyubiquitin Networks by Genomic, Interactome, and Sensor-Based Proteomic Analyses.
Legionella pneumophila regulates the activity of UBE2N by deamidase-mediated deubiquitination.
Legionella pneumophila inhibits immune signalling via MavC-mediated transglutaminase-induced ubiquitination of UBE2N.
OTUB1 non-catalytically stabilizes the E2 ubiquitin-conjugating enzyme UBE2E1 by preventing its autoubiquitination.
Genetic rescue of lineage-balanced blood cell production reveals a crucial role for STAT3 antiinflammatory activity in hematopoiesis.
ZUFSP Deubiquitylates K63-Linked Polyubiquitin Chains to Promote Genome Stability.
Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity.
A High-Throughput Screening Strategy for Development of RNF8-Ubc13 Protein-Protein Interaction Inhibitors.
HTLV-1 Tax Induces Formation of the Active Macromolecular IKK Complex by Generating Lys63- and Met1-Linked Hybrid Polyubiquitin Chains.
Human T-cell leukemia virus type 1 (HTLV-1) tax requires CADM1/TSLC1 for inactivation of the NF-κB inhibitor A20 and constitutive NF-κB signaling.
Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
A small-molecule inhibitor of UBE2N induces neuroblastoma cell death via activation of p53 and JNK pathways.
USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase.
STAT3 restrains RANK- and TLR4-mediated signalling by suppressing expression of the E2 ubiquitin-conjugating enzyme Ubc13.
A specific subset of E2 ubiquitin-conjugating enzymes regulate Parkin activation and mitophagy differently.
E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1.
Inhibition of proliferation and survival of diffuse large B-cell lymphoma cells by a small-molecule inhibitor of the ubiquitin-conjugating enzyme Ubc13-Uev1A.
Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress.
A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling.
The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage.
Regulation of hematopoiesis by the K63-specific ubiquitin-conjugating enzyme Ubc13.
Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly.
RNF8-dependent and RNF8-independent regulation of 53BP1 in response to DNA damage.
The human T-cell leukemia virus type 1 Tax oncoprotein requires the ubiquitin-conjugating enzyme Ubc13 for NF-kappaB activation.
Cutting Edge: Pivotal function of Ubc13 in thymocyte TCR signaling.
TIFA activates IkappaB kinase (IKK) by promoting oligomerization and ubiquitination of TRAF6.
Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB.
Ni J, Guan C, Liu H, Huang X, Yue J, Xiang H, Jiang Z, Tao Y, Cao W, Liu J, Wang Z, Wang Y, Wu X
Journal of immunology (Baltimore, Md. : 1950) 2021 May 15;206(10):2376-2385
Journal of immunology (Baltimore, Md. : 1950) 2021 May 15;206(10):2376-2385
Abraxas suppresses DNA end resection and limits break-induced replication by controlling SLX4/MUS81 chromatin loading in response to TOP1 inhibitor-induced DNA damage.
Wu X, Wang B
Nature communications 2021 Jul 16;12(1):4373
Nature communications 2021 Jul 16;12(1):4373
The lncRNA 'UCA1' modulates the response to chemotherapy of ovarian cancer through direct binding to miR-27a-5p and control of UBE2N levels.
Wambecke A, Ahmad M, Morice PM, Lambert B, Weiswald LB, Vernon M, Vigneron N, Abeilard E, Brotin E, Figeac M, Gauduchon P, Poulain L, Denoyelle C, Meryet-Figuiere M
Molecular oncology 2021 Dec;15(12):3659-3678
Molecular oncology 2021 Dec;15(12):3659-3678
Immune regulator LGP2 targets Ubc13/UBE2N to mediate widespread interference with K63 polyubiquitination and NF-κB activation.
Lenoir JJ, Parisien JP, Horvath CM
Cell reports 2021 Dec 28;37(13):110175
Cell reports 2021 Dec 28;37(13):110175
Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination.
Puvar K, Iyer S, Fu J, Kenny S, Negrón Terón KI, Luo ZQ, Brzovic PS, Klevit RE, Das C
Nature communications 2020 May 12;11(1):2365
Nature communications 2020 May 12;11(1):2365
Advanced Cataloging of Lysine-63 Polyubiquitin Networks by Genomic, Interactome, and Sensor-Based Proteomic Analyses.
Romero-Barrios N, Monachello D, Dolde U, Wong A, San Clemente H, Cayrel A, Johnson A, Lurin C, Vert G
The Plant cell 2020 Jan;32(1):123-138
The Plant cell 2020 Jan;32(1):123-138
Legionella pneumophila regulates the activity of UBE2N by deamidase-mediated deubiquitination.
Gan N, Guan H, Huang Y, Yu T, Fu J, Nakayasu ES, Puvar K, Das C, Wang D, Ouyang S, Luo ZQ
The EMBO journal 2020 Feb 17;39(4):e102806
The EMBO journal 2020 Feb 17;39(4):e102806
Legionella pneumophila inhibits immune signalling via MavC-mediated transglutaminase-induced ubiquitination of UBE2N.
Gan N, Nakayasu ES, Hollenbeck PJ, Luo ZQ
Nature microbiology 2019 Jan;4(1):134-143
Nature microbiology 2019 Jan;4(1):134-143
OTUB1 non-catalytically stabilizes the E2 ubiquitin-conjugating enzyme UBE2E1 by preventing its autoubiquitination.
Pasupala N, Morrow ME, Que LT, Malynn BA, Ma A, Wolberger C
The Journal of biological chemistry 2018 Nov 23;293(47):18285-18295
The Journal of biological chemistry 2018 Nov 23;293(47):18285-18295
Genetic rescue of lineage-balanced blood cell production reveals a crucial role for STAT3 antiinflammatory activity in hematopoiesis.
Zhang H, Li HS, Hillmer EJ, Zhao Y, Chrisikos TT, Hu H, Wu X, Thompson EJ, Clise-Dwyer K, Millerchip KA, Wei Y, Puebla-Osorio N, Kaushik S, Santos MA, Wang B, Garcia-Manero G, Wang J, Sun SC, Watowich SS
Proceedings of the National Academy of Sciences of the United States of America 2018 Mar 6;115(10):E2311-E2319
Proceedings of the National Academy of Sciences of the United States of America 2018 Mar 6;115(10):E2311-E2319
ZUFSP Deubiquitylates K63-Linked Polyubiquitin Chains to Promote Genome Stability.
Haahr P, Borgermann N, Guo X, Typas D, Achuthankutty D, Hoffmann S, Shearer R, Sixma TK, Mailand N
Molecular cell 2018 Apr 5;70(1):165-174.e6
Molecular cell 2018 Apr 5;70(1):165-174.e6
Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity.
Vujanovic M, Krietsch J, Raso MC, Terraneo N, Zellweger R, Schmid JA, Taglialatela A, Huang JW, Holland CL, Zwicky K, Herrador R, Jacobs H, Cortez D, Ciccia A, Penengo L, Lopes M
Molecular cell 2017 Sep 7;67(5):882-890.e5
Molecular cell 2017 Sep 7;67(5):882-890.e5
A High-Throughput Screening Strategy for Development of RNF8-Ubc13 Protein-Protein Interaction Inhibitors.
Weber E, Rothenaigner I, Brandner S, Hadian K, Schorpp K
SLAS discovery : advancing life sciences R & D 2017 Mar;22(3):316-323
SLAS discovery : advancing life sciences R & D 2017 Mar;22(3):316-323
HTLV-1 Tax Induces Formation of the Active Macromolecular IKK Complex by Generating Lys63- and Met1-Linked Hybrid Polyubiquitin Chains.
Shibata Y, Tokunaga F, Goto E, Komatsu G, Gohda J, Saeki Y, Tanaka K, Takahashi H, Sawasaki T, Inoue S, Oshiumi H, Seya T, Nakano H, Tanaka Y, Iwai K, Inoue JI
PLoS pathogens 2017 Jan;13(1):e1006162
PLoS pathogens 2017 Jan;13(1):e1006162
Human T-cell leukemia virus type 1 (HTLV-1) tax requires CADM1/TSLC1 for inactivation of the NF-κB inhibitor A20 and constitutive NF-κB signaling.
Pujari R, Hunte R, Thomas R, van der Weyden L, Rauch D, Ratner L, Nyborg JK, Ramos JC, Takai Y, Shembade N
PLoS pathogens 2015 Mar;11(3):e1004721
PLoS pathogens 2015 Mar;11(3):e1004721
Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Hodge CD, Edwards RA, Markin CJ, McDonald D, Pulvino M, Huen MS, Zhao J, Spyracopoulos L, Hendzel MJ, Glover JN
ACS chemical biology 2015 Jul 17;10(7):1718-28
ACS chemical biology 2015 Jul 17;10(7):1718-28
A small-molecule inhibitor of UBE2N induces neuroblastoma cell death via activation of p53 and JNK pathways.
Cheng J, Fan YH, Xu X, Zhang H, Dou J, Tang Y, Zhong X, Rojas Y, Yu Y, Zhao Y, Vasudevan SA, Zhang H, Nuchtern JG, Kim ES, Chen X, Lu F, Yang J
Cell death & disease 2014 Feb 20;5(2):e1079
Cell death & disease 2014 Feb 20;5(2):e1079
USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase.
Fan Y, Mao R, Yu Y, Liu S, Shi Z, Cheng J, Zhang H, An L, Zhao Y, Xu X, Chen Z, Kogiso M, Zhang D, Zhang H, Zhang P, Jung JU, Li X, Xu G, Yang J
The Journal of experimental medicine 2014 Feb 10;211(2):313-28
The Journal of experimental medicine 2014 Feb 10;211(2):313-28
STAT3 restrains RANK- and TLR4-mediated signalling by suppressing expression of the E2 ubiquitin-conjugating enzyme Ubc13.
Zhang H, Hu H, Greeley N, Jin J, Matthews AJ, Ohashi E, Caetano MS, Li HS, Wu X, Mandal PK, McMurray JS, Moghaddam SJ, Sun SC, Watowich SS
Nature communications 2014 Dec 15;5:5798
Nature communications 2014 Dec 15;5:5798
A specific subset of E2 ubiquitin-conjugating enzymes regulate Parkin activation and mitophagy differently.
Fiesel FC, Moussaud-Lamodière EL, Ando M, Springer W
Journal of cell science 2014 Aug 15;127(Pt 16):3488-504
Journal of cell science 2014 Aug 15;127(Pt 16):3488-504
E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1.
Wiener R, DiBello AT, Lombardi PM, Guzzo CM, Zhang X, Matunis MJ, Wolberger C
Nature structural & molecular biology 2013 Sep;20(9):1033-9
Nature structural & molecular biology 2013 Sep;20(9):1033-9
Inhibition of proliferation and survival of diffuse large B-cell lymphoma cells by a small-molecule inhibitor of the ubiquitin-conjugating enzyme Ubc13-Uev1A.
Pulvino M, Liang Y, Oleksyn D, DeRan M, Van Pelt E, Shapiro J, Sanz I, Chen L, Zhao J
Blood 2012 Aug 23;120(8):1668-77
Blood 2012 Aug 23;120(8):1668-77
Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress.
Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC, Livingston DM, Haracska L, Elledge SJ
Molecular cell 2012 Aug 10;47(3):396-409
Molecular cell 2012 Aug 10;47(3):396-409
A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling.
Cotta-Ramusino C, McDonald ER 3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ
Science (New York, N.Y.) 2011 Jun 10;332(6035):1313-7
Science (New York, N.Y.) 2011 Jun 10;332(6035):1313-7
The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage.
Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D
Cell 2009 Feb 6;136(3):420-34
Cell 2009 Feb 6;136(3):420-34
Regulation of hematopoiesis by the K63-specific ubiquitin-conjugating enzyme Ubc13.
Wu X, Yamamoto M, Akira S, Sun SC
Proceedings of the National Academy of Sciences of the United States of America 2009 Dec 8;106(49):20836-41
Proceedings of the National Academy of Sciences of the United States of America 2009 Dec 8;106(49):20836-41
Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly.
Huen MS, Huang J, Yuan J, Yamamoto M, Akira S, Ashley C, Xiao W, Chen J
Molecular and cellular biology 2008 Oct;28(19):6104-12
Molecular and cellular biology 2008 Oct;28(19):6104-12
RNF8-dependent and RNF8-independent regulation of 53BP1 in response to DNA damage.
Sakasai R, Tibbetts R
The Journal of biological chemistry 2008 May 16;283(20):13549-55
The Journal of biological chemistry 2008 May 16;283(20):13549-55
The human T-cell leukemia virus type 1 Tax oncoprotein requires the ubiquitin-conjugating enzyme Ubc13 for NF-kappaB activation.
Shembade N, Harhaj NS, Yamamoto M, Akira S, Harhaj EW
Journal of virology 2007 Dec;81(24):13735-42
Journal of virology 2007 Dec;81(24):13735-42
Cutting Edge: Pivotal function of Ubc13 in thymocyte TCR signaling.
Yamamoto M, Sato S, Saitoh T, Sakurai H, Uematsu S, Kawai T, Ishii KJ, Takeuchi O, Akira S
Journal of immunology (Baltimore, Md. : 1950) 2006 Dec 1;177(11):7520-4
Journal of immunology (Baltimore, Md. : 1950) 2006 Dec 1;177(11):7520-4
TIFA activates IkappaB kinase (IKK) by promoting oligomerization and ubiquitination of TRAF6.
Ea CK, Sun L, Inoue J, Chen ZJ
Proceedings of the National Academy of Sciences of the United States of America 2004 Oct 26;101(43):15318-23
Proceedings of the National Academy of Sciences of the United States of America 2004 Oct 26;101(43):15318-23
Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB.
Habelhah H, Takahashi S, Cho SG, Kadoya T, Watanabe T, Ronai Z
The EMBO journal 2004 Jan 28;23(2):322-32
The EMBO journal 2004 Jan 28;23(2):322-32
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
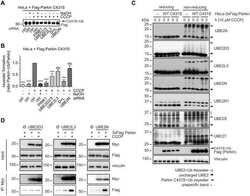
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
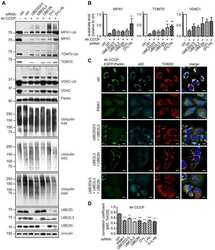
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
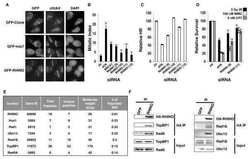
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
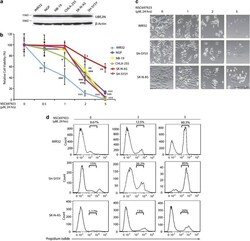
- Experimental details
- Figure 1 NSC697923 shows cytotoxic effect on NB cell lines. ( a ) A panel of six NB cell lines were lysed, subjected to SDS-PAGE and immunoblotted with UBE2N antibodies. Beta-actin was detected as a loading control. ( b ) Six NB cell lines were treated with the indicated concentrations of NSC697923 for 24 h. Cell viability was then measured by adding 10 mu l of CCK-8 and reading the absorbance at 450 nm. Data were represented as mean+-S.D. P -values
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
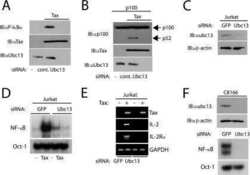
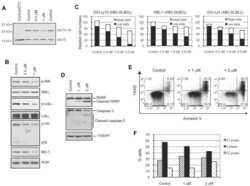
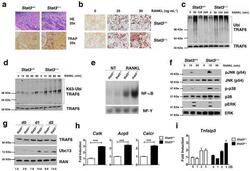
- Figure 1 STAT3 suppresses osteoclastogenesis and RANK signal transduction (a) Femoral sections from Stat3 -deficient ( Stat3 delta/delta ) and -sufficient ( Stat3 +/+ ) mice, 8 weeks. ( Upper ) Hematoxylin and eosin (HE); ( Lower ) TRAP staining (arrows). Scale bar, 100 M. ( b to i ) Bone marrow cells from Stat3 +/+ and Stat3 delta/delta mice were cultured in M-CSF-containing medium for 4 d to generate bone marrow-derived macrophages. ( b ) 2x10 5 cells were cultured in M-CSF and RANKL (0, 25 or 50 ng mL -1 ) for an additional 4 d. Osteoclasts detected by TRAP staining (pink). Scale bar, 100 M ( c to f ) Macrophages were starved of exogenous growth factor overnight and stimulated with RANKL (100 ng mL -1 ) or left untreated, as indicated. ( c and d ) TRAF6 ubiquitination was evaluated -/+ RANKL stimulation by immunoprecipitation and immunoblotting for total ubiquitin ( c ) or K63-specific linkage ( d ). Lower panels: total TRAF6. ( e ) Nuclear NF-kappaB DNA binding activity was measured -/+ RANKL stimulation (15 min) by EMSA. Loading control: NF-Y EMSA. ( f ) MAPK activation was measured -/+ RANKL stimulation by immunoblotting whole cell lysates with antibodies specific for phosphorylated protein (pJNK, p-p38 or pERK). Total protein amounts served as loading controls (JNK, p38 or ERK). ( g ) Macrophages were cultured in M-CSF and RANKL (50 ng mL -1 ). TRAF6 and Ubc13 amounts were determined by immunoblotting. Loading control: RAN. The relative level of Ubc13 is expressed as a
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
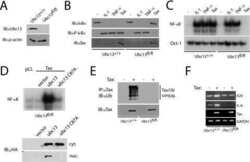
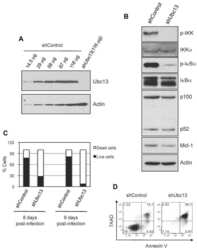
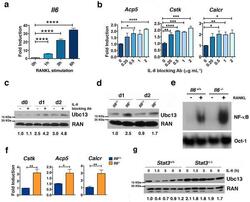
- Figure 3 Autocrine IL-6 is required to repress Ubc13 expression a ) Il6 mRNA expression was measured in bone marrow-derived macrophages by qPCR following RANKL (50 ng mL -1 ) stimulation, as indicated. ( b and c ) Osteoclast gene expression (b) and Ubc13 amounts (c) were determined in bone marrow-derived macrophages following culture in M-CSF- and RANKL-containing medium without (-) or with (+) IL-6 blocking antibody by qPCR or immunoblotting, respectively. (d to f) Ubc13 amounts, nuclear NF-kappaB DNA binding activity and osteoclast gene expression were determined in Il6 +/+ and Il6 -/- bone marrow-derived macrophages following culture in M-CSF- and RANKL-containing medium for 1 or 2 d ( d ), 1 d ( e ), or 3 d ( f ) by immunoblotting, EMSA or qPCR, respectively. (g) Ubc13 expression was evaluated in bone marrow-derived macrophages from Stat3 -sufficient ( Stat3 +/+ ) or hematopoietic Stat3 -deficient ( Stat3 delta/delta ) mice by immunoblotting following IL-6 stimulation, as indicated. ( c , d and g ) Relative Ubc13 expression vs. untreated ( c ), IL-6-( d ) or STAT3-sufficient control ( g ) was calculated as indicated in Fig. 1g and displayed below bottom panels. ( a and f ) Results represent mean values of 3 independent experiments. Error bars indicate SEM. ( a and b ) Two-way Anova with Bonferroni multiple comparison used; ( f ) t test used. *, p
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
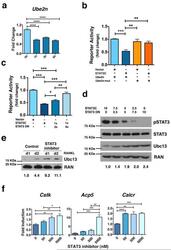
- Figure 4 STAT3 transcriptional activity is required to repress Ube2n expression ( a ) Ube2n mRNA was measured in bone marrow-derived macrophages from Stat3 -sufficient mice following treatment with IL-6 as indicated, using qPCR. (b and c) Ube2n promoter activity was measured by reporter assays in HEK293T cells transfected with pGL3- Ube2n ( Ube2n ), pGL3- Ube2n -mut ( Ube2n -mut; containing a mutated STATx site), pMX vector (vector), pMX-STAT3C (STAT3C), pMX-STAT3 DN (STAT3 DN) and pTK-Renilla, as indicated. pMX-STAT3 DN and pMX-STAT3C were included in some samples in a 3:1 or 6:1 ratio, as shown. (d) Stat3 -/- MEFs were transfected with pMX-STAT3C, pMX-STAT3 DN or both, at the ratios indicated. The expression level of pSTAT3, total STAT3 and Ubc13 was detected by immunoblotting. Ran served as loading control . (e and f) Bone marrow-derived macrophages from Stat3 -sufficient mice were cultured in M-CSF- and RANKL-containing medium in the absence or presence of the STAT3 inhibitor PM-73G, as indicated. Ubc13 amounts were detected by immunoblotting ( e ); osteoclast gene expression was measured by qPCR ( f ). ( d and e ) Relative Ubc13 expression vs. STAT3C transfection alone ( d ) or untreated cells on 1 d ( e ) was calculated as indicated in Fig. 1g and displayed below bottom panels. (a, b, c and f) Data represent mean values of 3 independent experiments. Error bars indicate SEM. Two-way Anova with Bonferroni multiple comparison used. *, p
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
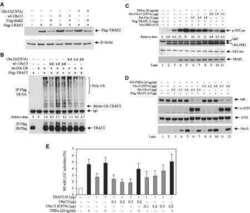
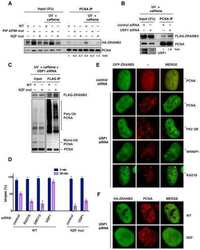
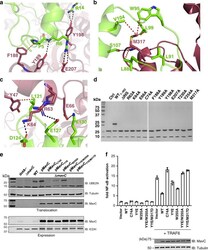
- Fig. 5 Binding interface of MavC with Ube2N. a-c Detailed view of interactions in MavC regions 1, 2, and 3 with Ube2N. Key residues are labeled and represented as stick models. MavC is depicted in burgundy and Ube2N in green. Hydrogen-bonding interactions are given in black and hydrophobic interactions in red. d Comparison of the ubiquitinating activity of wild-type MavC versus mutant proteins using Ube2N-SS-Ub as the substrate. Reactions were subjected to SDS-PAGE and visualized with Coomassie Blue. A control reaction without MavC is included. e MavC-mediated ubiquitination of Ube2N during L.p infection. Cells infected with the indicated L. pneumophila strains were lysed with 0.2% saponin and lysate separated by SDS-PAGE, probed by immunoblotting with antibodies specific for Ube2N (upper panel) and MavC (lower panel). respectively. f The effects of MavC and its mutants on NFkappaB activation. HEK293T cells were transfected with plasmids expressing a luciferase reporter responsive to NF-kappaB and Flag-MavC or its mutant. At the same time, a plasmid expressing Renilla luciferase used as an internal control and stimulator TRAF6 were co-transfected. NF-kappaB activity was determined by dual luciferase assay. The expression of MavC and its mutants was probed in lysates of transfected cell while tubulin was detected as a loading control. Three independent experiments were done with similar results. Error bars indicate standard error of the mean (SEM).
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- 5 Fig. UCA1 inhibition releases miR-27a-5p which downregulates UBE2N and induces BIM expression. (A) Relative transcript distribution of UCA1 short and long isoforms (RT-qPCR) in nuclear and cytoplasmic compartments in OAW42-R cells normalized to total cellular transcript levels. NEAT1 was used as a control for nuclear transcript. (B) Venn diagram of overlapping miRNAs between miRNAs reported/predicted to bind UCA1 (left) and miRNAs whose seed is enriched in DEGs (right) after UCA1 downregulation in OAW42-R cells. (C) Schematic representation of short isoform of UCA1 and miR-27a-5p sequence complementarity as predicted by RNA22 v2 software. (D) Fold enrichment of UCA1 after Biotin-miR-27a-5p pull-down relative to Biotin-Cel-miR-67 pull-down. Data are expressed as mean +- SEM from 3 experiments. (E, G and H) BIM, PUMA, or UBE2N (unmodified and ubiquitinated) protein levels (western blot), quantified relative to actin loading control, representative or mean +- SEM of 3 experiments, after indicated siRNAs transfection in OAW42-R cells. (F) Pictures from cell layer. Data are representative of 3 experiments
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
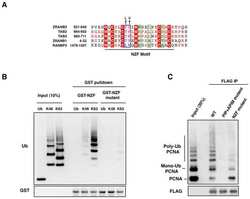
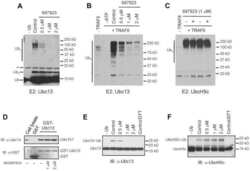
- Figure 1 E2 enzymes stimulate OTUB1 DUB activity ( a ) FRET-based assay for cleavage of internally quenched fluorescent K48 Ub 2 (400 nM) by OTUB1 (30 nM) in the absence and presence of 11 different E2 enzymes (5 uM). ( b ) The rates of K48 Ub 2 (400 nM) cleavage by OTUB1 (30 nM) are plotted as a function of the log of the concentration of UBCH5B. The data were analyzed and fit using a ""log(agonist) vs. response"" model to determine EC 50 . Each rate was measured in triplicate, and error bars represent the SEM for each measurement. ( c ) Coomassie stained gel showing K48 Ub 2 (15 uM) cleavage by OTUB1 (0.5 uM) in the presence and absence of UBCH5B (25 uM). ( d ) Steady-state kinetic saturation curve for cleavage of K48 Ub 2 by OTUB1 (30 nM) in the presence (red) and absence (blue) of UBCH5B (10 uM). Each rate was measured in triplicate, and error bars represent the SEM for each measurement.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
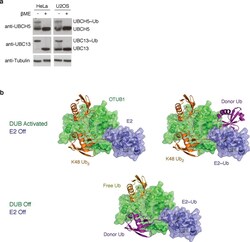
- Figure 5 E2 enzymes exist in both charged and uncharged states in vivo ( a ) Western blot of cell lysates from HeLa and U2OS cells prepared at acidic pH (+- beta-ME) and probed with anti-UBC13 and anti-UBCH5 antibodies exhibit both charged and uncharged forms of the E2 enzymes. ( b ) Depiction of three possible states of OTUB1-E2 complexes. Top: OTUB1 (green) bound to K48 Ub 2 (orange) and E2 (blue) or E2 (blue)~Ub (purple) thioester. Bottom: OTUB1 bound to free Ub (yellow) and E2~Ub thioester. The free ubiquitin (yellow) and the ubiquitin (purple) conjugated to the E2 bind in place of K48 Ub 2 .
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot ELISA
ELISA Other assay
Other assay