Antibody data
- Antibody Data
- Antigen structure
- References [6]
- Comments [0]
- Validations
- Immunocytochemistry [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA005848 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA005848, RRID:AB_1078768
- Product name
- Anti-EXD2
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human EXD2, Gene description: exonuclease 3'-5' domain containing 2, Alternative Gene Names: C14orf114, EXDL2, FLJ10738, Validated applications: ICC, IHC, WB, Uniprot ID: Q9NVH0, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human, Mouse
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.8 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references Active mRNA degradation by EXD2 nuclease elicits recovery of transcription after genotoxic stress
Pathway choice in the alternative telomere lengthening in neoplasia is dictated by replication fork processing mediated by EXD2’s nuclease activity
Spatiotemporally-resolved mapping of RNA binding proteins via functional proximity labeling reveals a mitochondrial mRNA anchor promoting stress recovery
EXD2 Protects Stressed Replication Forks and Is Required for Cell Viability in the Absence of BRCA1/2
The mitochondrial outer-membrane location of the EXD2 exonuclease contradicts its direct role in nuclear DNA repair
DNA Double-Strand Break Resection Occurs during Non-homologous End Joining in G1 but Is Distinct from Resection during Homologous Recombination
Sandoz J, Cigrang M, Zachayus A, Catez P, Donnio L, Elly C, Nieminuszczy J, Berico P, Braun C, Alekseev S, Egly J, Niedzwiedz W, Giglia-Mari G, Compe E, Coin F
Nature Communications 2023;14(1)
Nature Communications 2023;14(1)
Pathway choice in the alternative telomere lengthening in neoplasia is dictated by replication fork processing mediated by EXD2’s nuclease activity
Broderick R, Cherdyntseva V, Nieminuszczy J, Dragona E, Kyriakaki M, Evmorfopoulou T, Gagos S, Niedzwiedz W
Nature Communications 2023;14(1)
Nature Communications 2023;14(1)
Spatiotemporally-resolved mapping of RNA binding proteins via functional proximity labeling reveals a mitochondrial mRNA anchor promoting stress recovery
Qin W, Myers S, Carey D, Carr S, Ting A
Nature Communications 2021;12(1)
Nature Communications 2021;12(1)
EXD2 Protects Stressed Replication Forks and Is Required for Cell Viability in the Absence of BRCA1/2
Nieminuszczy J, Broderick R, Bellani M, Smethurst E, Schwab R, Cherdyntseva V, Evmorfopoulou T, Lin Y, Minczuk M, Pasero P, Gagos S, Seidman M, Niedzwiedz W
Molecular Cell 2019;75(3):605-619.e6
Molecular Cell 2019;75(3):605-619.e6
The mitochondrial outer-membrane location of the EXD2 exonuclease contradicts its direct role in nuclear DNA repair
Hensen F, Moretton A, van Esveld S, Farge G, Spelbrink J
Scientific Reports 2018;8(1)
Scientific Reports 2018;8(1)
DNA Double-Strand Break Resection Occurs during Non-homologous End Joining in G1 but Is Distinct from Resection during Homologous Recombination
Biehs R, Steinlage M, Barton O, Juhász S, Künzel J, Spies J, Shibata A, Jeggo P, Löbrich M
Molecular Cell 2017;65(4):671-684.e5
Molecular Cell 2017;65(4):671-684.e5
No comments: Submit comment
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
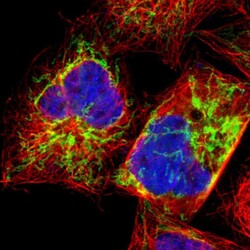
- Experimental details
- Immunofluorescent staining of human cell line A-431 shows localization to mitochondria.
- Sample type
- Human
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry