Antibody data
- Antibody Data
- Antigen structure
- References [12]
- Comments [0]
- Validations
- Western blot [2]
- Immunocytochemistry [1]
- Immunohistochemistry [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA003890 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA003890, RRID:AB_1078103
- Product name
- Anti-ADAR
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human ADAR, Gene description: adenosine deaminase, RNA-specific, Alternative Gene Names: ADAR1, G1P1, IFI4, Validated applications: WB, IHC, ICC, Uniprot ID: P55265, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.1 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references ADAR1 promotes cisplatin resistance in intrahepatic cholangiocarcinoma by regulating BRCA2 expression through A‐to‐I editing manner
Extracellular vesicle‑mediated RNA editing may underlie the heterogeneity and spread of hepatocellular carcinoma in human tissue and in vitro.
ADAR1 downregulation by autophagy drives senescence independently of RNA editing by enhancing p16INK4a levels
Heterogeneous RNA editing and influence of ADAR2 on mesothelioma chemoresistance and the tumor microenvironment
Double-Stranded RNA Structural Elements Holding the Key to Translational Regulation in Cancer: The Case of Editing in RNA-Binding Motif Protein 8A
How asbestos drives the tissue towards tumors: YAP activation, macrophage and mesothelial precursor recruitment, RNA editing, and somatic mutations
Decreased A-to-I RNA editing as a source of keratinocytes' dsRNA in psoriasis.
ADAR1 deletion induces NFκB and interferon signaling dependent liver inflammation and fibrosis.
A novel immune resistance mechanism of melanoma cells controlled by the ADAR1 enzyme
MicroRNA Editing Facilitates Immune Elimination of HCMV Infected Cells
ADAR-Related Activation of Adenosine-to-Inosine RNA Editing During Regeneration
Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature
Liu Q, Huang C, Chen S, Zhu Y, Huang X, Zhao G, Xu Q, Shi Y, Li W, Wang R, Yin X
Cell Proliferation 2024;57(10)
Cell Proliferation 2024;57(10)
Extracellular vesicle‑mediated RNA editing may underlie the heterogeneity and spread of hepatocellular carcinoma in human tissue and in vitro.
Shibata C, Otsuka M, Shimizu T, Seimiya T, Kishikawa T, Aoki T, Fujishiro M
Oncology reports 2023 Nov;50(5)
Oncology reports 2023 Nov;50(5)
ADAR1 downregulation by autophagy drives senescence independently of RNA editing by enhancing p16INK4a levels
Hao X, Shiromoto Y, Sakurai M, Towers M, Zhang Q, Wu S, Havas A, Wang L, Berger S, Adams P, Tian B, Nishikura K, Kossenkov A, Liu P, Zhang R
Nature Cell Biology 2022;24(8):1202-1210
Nature Cell Biology 2022;24(8):1202-1210
Heterogeneous RNA editing and influence of ADAR2 on mesothelioma chemoresistance and the tumor microenvironment
Hariharan A, Qi W, Rehrauer H, Wu L, Ronner M, Wipplinger M, Kresoja‐Rakic J, Sun S, Oton‐Gonzalez L, Sculco M, Serre‐Beinier V, Meiller C, Blanquart C, Fonteneau J, Vrugt B, Rüschoff J, Opitz I, Jean D, de Perrot M, Felley‐Bosco E
Molecular Oncology 2022;16(22):3949-3974
Molecular Oncology 2022;16(22):3949-3974
Double-Stranded RNA Structural Elements Holding the Key to Translational Regulation in Cancer: The Case of Editing in RNA-Binding Motif Protein 8A
Abukar A, Wipplinger M, Hariharan A, Sun S, Ronner M, Sculco M, Okonska A, Kresoja-Rakic J, Rehrauer H, Qi W, Beusechem V, Felley-Bosco E
Cells 2021;10(12):3543
Cells 2021;10(12):3543
How asbestos drives the tissue towards tumors: YAP activation, macrophage and mesothelial precursor recruitment, RNA editing, and somatic mutations
Rehrauer H, Wu L, Blum W, Pecze L, Henzi T, Serre-Beinier V, Aquino C, Vrugt B, de Perrot M, Schwaller B, Felley-Bosco E
Oncogene 2018;37(20):2645-2659
Oncogene 2018;37(20):2645-2659
Decreased A-to-I RNA editing as a source of keratinocytes' dsRNA in psoriasis.
Shallev L, Kopel E, Feiglin A, Leichner GS, Avni D, Sidi Y, Eisenberg E, Barzilai A, Levanon EY, Greenberger S
RNA (New York, N.Y.) 2018 Jun;24(6):828-840
RNA (New York, N.Y.) 2018 Jun;24(6):828-840
ADAR1 deletion induces NFκB and interferon signaling dependent liver inflammation and fibrosis.
Ben-Shoshan SO, Kagan P, Sultan M, Barabash Z, Dor C, Jacob-Hirsch J, Harmelin A, Pappo O, Marcu-Malina V, Ben-Ari Z, Amariglio N, Rechavi G, Goldstein I, Safran M
RNA biology 2017 May 4;14(5):587-602
RNA biology 2017 May 4;14(5):587-602
A novel immune resistance mechanism of melanoma cells controlled by the ADAR1 enzyme
Galore-Haskel G, Nemlich Y, Greenberg E, Ashkenazi S, Hakim M, Itzhaki O, Shoshani N, Shapira-Fromer R, Ben-Ami E, Ofek E, Anafi L, Besser M, Schachter J, Markel G
Oncotarget 2015;6(30):28999-29015
Oncotarget 2015;6(30):28999-29015
MicroRNA Editing Facilitates Immune Elimination of HCMV Infected Cells
Gao S, Nachmani D, Zimmermann A, Oiknine Djian E, Weisblum Y, Livneh Y, Khanh Le V, Galun E, Horejsi V, Isakov O, Shomron N, Wolf D, Hengel H, Mandelboim O
PLoS Pathogens 2014;10(2):e1003963
PLoS Pathogens 2014;10(2):e1003963
ADAR-Related Activation of Adenosine-to-Inosine RNA Editing During Regeneration
Witman N, Behm M, Öhman M, Morrison J
Stem Cells and Development 2013;22(16):2254-2267
Stem Cells and Development 2013;22(16):2254-2267
Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature
Rice G, Kasher P, Forte G, Mannion N, Greenwood S, Szynkiewicz M, Dickerson J, Bhaskar S, Zampini M, Briggs T, Jenkinson E, Bacino C, Battini R, Bertini E, Brogan P, Brueton L, Carpanelli M, De Laet C, de Lonlay P, del Toro M, Desguerre I, Fazzi E, Garcia-Cazorla À, Heiberg A, Kawaguchi M, Kumar R, Lin J, Lourenco C, Male A, Marques W, Mignot C, Olivieri I, Orcesi S, Prabhakar P, Rasmussen M, Robinson R, Rozenberg F, Schmidt J, Steindl K, Tan T, van der Merwe W, Vanderver A, Vassallo G, Wakeling E, Wassmer E, Whittaker E, Livingston J, Lebon P, Suzuki T, McLaughlin P, Keegan L, O'Connell M, Lovell S, Crow Y
Nature Genetics 2012;44(11):1243-1248
Nature Genetics 2012;44(11):1243-1248
No comments: Submit comment
Enhanced validation
Enhanced validation
- Submitted by
- klas2
- Enhanced method
- Genetic validation
- Main image
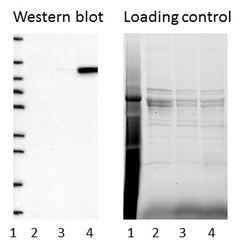
- Experimental details
- Western blot of cell lysate from U-2 OS cells transfected with either siRNA targeting ADAR or control siRNA. Lane 1: Marker (250, 130, 95, 72, 55, 36, 28, 17, 10) Lane 2: Cell lysate from U-2OS cells transfected with siRNA targeting ADAR Lane 3: N/A Lane 4: Cell lysate from U-2OS cells transfected with control siRNA Right image, lane 1-4: loading control
- Sample type
- U-2 OS
- Primary Ab dilution
- 1:196
- Conjugate
- Horseradish Peroxidase
- Secondary Ab
- Secondary Ab
- Secondary Ab dilution
- 1:3000
- Knockdown/Genetic Approaches Application
- Western blot
Enhanced validation
- Submitted by
- Atlas Antibodies (provider)
- Enhanced method
- Genetic validation
- Main image
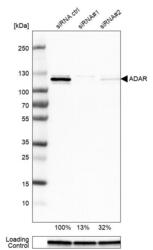
- Experimental details
- Western blot analysis in U-251MG cells transfected with control siRNA, target specific siRNA probe #1 and #2, using Anti-ADAR antibody. Remaining relative intensity is presented. Loading control: Anti-GAPDH.
- Sample type
- Human
- Protocol
- Protocol
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
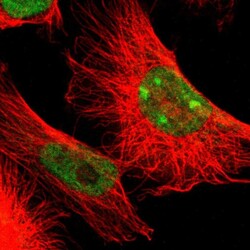
- Experimental details
- Immunofluorescent staining of human cell line U-251 MG shows localization to nucleoplasm & nucleoli.
- Sample type
- Human
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Enhanced method
- Orthogonal validation
- Main image
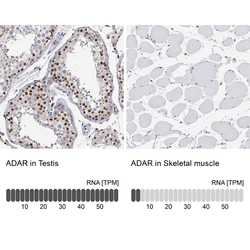
- Experimental details
- Immunohistochemistry analysis in human testis and skeletal muscle tissues using HPA003890 antibody. Corresponding ADAR RNA-seq data are presented for the same tissues.
- Sample type
- Human
- Protocol
- Protocol
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry