Antibody data
- Antibody Data
- Antigen structure
- References [4]
- Comments [0]
- Validations
- Immunocytochemistry [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA020949 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA020949, RRID:AB_1857223
- Product name
- Anti-ZMYND8
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human ZMYND8, Gene description: zinc finger, MYND-type containing 8, Alternative Gene Names: PRKCBP1, RACK7, Validated applications: IHC, ICC, Uniprot ID: Q9ULU4, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.2 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references ZMYND8 mediated liquid condensates spatiotemporally decommission the latent super-enhancers during macrophage polarization
Functional antagonism of chromatin modulators regulates epithelial-mesenchymal transition.
The Chromatin Reader ZMYND8 Regulates Igh Enhancers to Promote Immunoglobulin Class Switch Recombination
Selective Recognition of H3.1K36 Dimethylation/H4K16 Acetylation Facilitates the Regulation of All-trans-retinoic Acid (ATRA)-responsive Genes by Putative Chromatin Reader ZMYND8
Jia P, Li X, Wang X, Yao L, Xu Y, Hu Y, Xu W, He Z, Zhao Q, Deng Y, Zang Y, Zhang M, Zhang Y, Qin J, Lu W
Nature Communications 2021;12(1)
Nature Communications 2021;12(1)
Functional antagonism of chromatin modulators regulates epithelial-mesenchymal transition.
Serresi M, Kertalli S, Li L, Schmitt MJ, Dramaretska Y, Wierikx J, Hulsman D, Gargiulo G
Science advances 2021 Feb;7(9)
Science advances 2021 Feb;7(9)
The Chromatin Reader ZMYND8 Regulates Igh Enhancers to Promote Immunoglobulin Class Switch Recombination
Delgado-Benito V, Rosen D, Wang Q, Gazumyan A, Pai J, Oliveira T, Sundaravinayagam D, Zhang W, Andreani M, Keller L, Kieffer-Kwon K, Pękowska A, Jung S, Driesner M, Subbotin R, Casellas R, Chait B, Nussenzweig M, Di Virgilio M
Molecular Cell 2018;72(4):636-649.e8
Molecular Cell 2018;72(4):636-649.e8
Selective Recognition of H3.1K36 Dimethylation/H4K16 Acetylation Facilitates the Regulation of All-trans-retinoic Acid (ATRA)-responsive Genes by Putative Chromatin Reader ZMYND8
Adhikary S, Sanyal S, Basu M, Sengupta I, Sen S, Srivastava D, Roy S, Das C
Journal of Biological Chemistry 2016;291(6):2664-2681
Journal of Biological Chemistry 2016;291(6):2664-2681
No comments: Submit comment
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
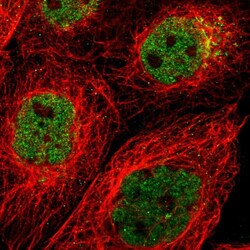
- Experimental details
- Immunofluorescent staining of human cell line A-431 shows localization to nucleoplasm & the Golgi apparatus.
- Sample type
- Human
 Explore
Explore Validate
Validate Learn
Learn Immunocytochemistry
Immunocytochemistry Immunohistochemistry
Immunohistochemistry