Antibody data
- Antibody Data
- Antigen structure
- References [7]
- Comments [0]
- Validations
- Immunocytochemistry [2]
- Immunohistochemistry [2]
- Other assay [5]
Submit
Validation data
Reference
Comment
Report error
- Product number
- MA5-16312 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- TET1 Monoclonal Antibody (GT1462)
- Antibody type
- Monoclonal
- Antigen
- Recombinant full-length protein
- Description
- Recommended positive controls: 293T, DDDDK-human TET1-transfected 293T, NT2D1, NT2D1 nuclear extract. Store product as a concentrated solution. Centrifuge briefly prior to opening the vial.
- Reactivity
- Human, Mouse
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- GT1462
- Vial size
- 100 μL
- Concentration
- 1 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references PROSER1 mediates TET2 O-GlcNAcylation to regulate DNA demethylation on UTX-dependent enhancers and CpG islands.
Ten-Eleven Translocation 1 Promotes Malignant Progression of Cholangiocarcinoma With Wild-Type Isocitrate Dehydrogenase 1.
Ligand-induced gene activation is associated with oxidative genome damage whose repair is required for transcription.
Integrin α6 signaling induces STAT3-TET3-mediated hydroxymethylation of genes critical for maintenance of glioma stem cells.
LncRNA NUTM2A-AS1 positively modulates TET1 and HIF-1A to enhance gastric cancer tumorigenesis and drug resistance by sponging miR-376a.
Vitamin C supports conversion of human γδ T cells into FOXP3-expressing regulatory cells by epigenetic regulation.
Hemodynamic disturbed flow induces differential DNA methylation of endothelial Kruppel-Like Factor 4 promoter in vitro and in vivo.
Wang X, Rosikiewicz W, Sedkov Y, Martinez T, Hansen BS, Schreiner P, Christensen J, Xu B, Pruett-Miller SM, Helin K, Herz HM
Life science alliance 2022 Jan;5(1)
Life science alliance 2022 Jan;5(1)
Ten-Eleven Translocation 1 Promotes Malignant Progression of Cholangiocarcinoma With Wild-Type Isocitrate Dehydrogenase 1.
Bai X, Zhang H, Zhou Y, Nagaoka K, Meng J, Ji C, Liu D, Dong X, Cao K, Mulla J, Cheng Z, Mueller W, Bay A, Hildebrand G, Lu S, Wallace J, Wands JR, Sun B, Huang CK
Hepatology (Baltimore, Md.) 2021 May;73(5):1747-1763
Hepatology (Baltimore, Md.) 2021 May;73(5):1747-1763
Ligand-induced gene activation is associated with oxidative genome damage whose repair is required for transcription.
Sengupta S, Wang H, Yang C, Szczesny B, Hegde ML, Mitra S
Proceedings of the National Academy of Sciences of the United States of America 2020 Sep 8;117(36):22183-22192
Proceedings of the National Academy of Sciences of the United States of America 2020 Sep 8;117(36):22183-22192
Integrin α6 signaling induces STAT3-TET3-mediated hydroxymethylation of genes critical for maintenance of glioma stem cells.
Herrmann A, Lahtz C, Song J, Aftabizadeh M, Cherryholmes GA, Xin H, Adamus T, Lee H, Grunert D, Armstrong B, Chu P, Brown C, Lim M, Forman S, Yu H
Oncogene 2020 Mar;39(10):2156-2169
Oncogene 2020 Mar;39(10):2156-2169
LncRNA NUTM2A-AS1 positively modulates TET1 and HIF-1A to enhance gastric cancer tumorigenesis and drug resistance by sponging miR-376a.
Wang J, Yu Z, Wang J, Shen Y, Qiu J, Zhuang Z
Cancer medicine 2020 Dec;9(24):9499-9510
Cancer medicine 2020 Dec;9(24):9499-9510
Vitamin C supports conversion of human γδ T cells into FOXP3-expressing regulatory cells by epigenetic regulation.
Kouakanou L, Peters C, Sun Q, Floess S, Bhat J, Huehn J, Kabelitz D
Scientific reports 2020 Apr 16;10(1):6550
Scientific reports 2020 Apr 16;10(1):6550
Hemodynamic disturbed flow induces differential DNA methylation of endothelial Kruppel-Like Factor 4 promoter in vitro and in vivo.
Jiang YZ, Jiménez JM, Ou K, McCormick ME, Zhang LD, Davies PF
Circulation research 2014 Jun 20;115(1):32-43
Circulation research 2014 Jun 20;115(1):32-43
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
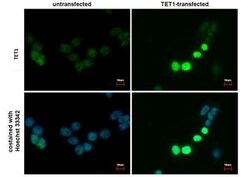
- Experimental details
- TET1 Monoclonal Antibody (GT1462) detects TET1 protein at nucleus by immunofluorescent analysis. Sample: TET1-transfected (right) or untransfected (left) 293T cells were fixed in 4% paraformaldehyde for 15 min. Green: TET1 protein stained by TET1 antibody (Product # MA5-16312) diluted at 1:1,000. Blue: Hoechst 33342 staining. Scale bar = 10 µm.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
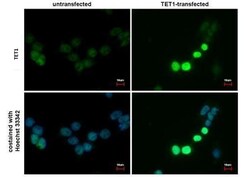
- Experimental details
- TET1 Monoclonal Antibody (GT1462) detects TET1 protein at nucleus by immunofluorescent analysis. Sample: TET1-transfected (right) or untransfected (left) 293T cells were fixed in 4% paraformaldehyde for 15 min. Green: TET1 protein stained by TET1 antibody (Product # MA5-16312) diluted at 1:1,000. Blue: Hoechst 33342 staining. Scale bar = 10 µm.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
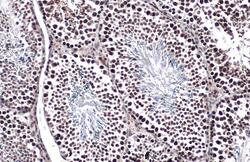
- Experimental details
- TET1 Monoclonal Antibody (GT1462) detects TET1 protein at nucleus by immunohistochemical analysis. Sample: Paraffin-embedded mouse testis. TET1 stained by TET1 Monoclonal Antibody (GT1462) (Product # MA5-16312) diluted at 1:500. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
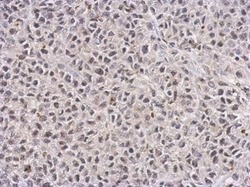
- Experimental details
- TET1 Monoclonal Antibody (GT1462) detects TET1 protein at nucleus on HeLa xenograft by immunohistochemical analysis. Sample: Paraffin-embedded HeLa xenograft. TET1 Monoclonal Antibody (GT1462) (Product # MA5-16312) dilution: 1:100. Antigen Retrieval: EDTA based buffer, pH 8.0, 15 min.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
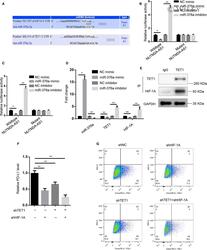
- 5 Figure miR-376a directly targeted TET1 and HIF1A. (A) Bioinformatics prediction (TargetScan database) of putative binding site at 3'-UTRs of TET1 and HIF-1A by miR-376a. (B and C) Luciferase activity was measured in HGC-27 cells transfected with HIF-1A or TET1 3'-UTR-fused pGL3 vector plus NC mimic, miR-376a mimic, NC inhibitor, miR-376a inhibitor. * p < 0.05; ** p < 0.01. (D) RT-qPCR showed miR-376a, TET1, HIF-1A expression levels in HGC-27 cells transfected with NC mimic, miR-376a mimic, NC inhibitor, miR-376a inhibitor. * p < 0.05; ** p < 0.01. (E) Co-immunoprecipitation showed interaction of TET1 with HIF-1A in HGC-27 cells. IgG served as negative control. (F) RT-qPCR showed PD-L1 mRNA levels in HGC-27 cells transfected with shNC, shHIF-1A, shTET1, shHIF-1A plus shTET1. ** p < 0.01. (G) Flow cytometry analysis showed PD-L1 protein levels in HGC-27 cells transfected with shNC, shHIF-1A, shTET1, shHIF-1A plus shTET1
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
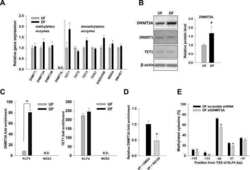
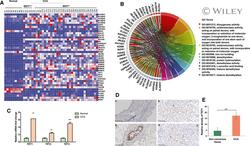
- 1 Fig. TET1 expression is elevated in patients with CCA. (A) Heat map for the expression of 2-OG-dependent enzymes was generated using TCGA data, which include 9 normal samples, 6 CCA tumors with IDH1 mutation, and 30 CCA tumors with WT IDH1. (B) The association of molecular signaling pathways and functions with alpha-KG-dependent enzymes. (C) Relative TET1, TET2, and TET3 mRNA expression levels were analyzed in human normal bile ducts (n = 9) and patients with CCA (n = 36) using TCGA data. (D) Representative TET1 immunohistochemical images are shown in noncancerous bile duct (a) and CCA samples (b-d). (E) Semiquantification of TET1 immunohistochemical staining data was calculated using Image J software. ** P < 0.01, *** P < 0.001 compared with relevant control.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
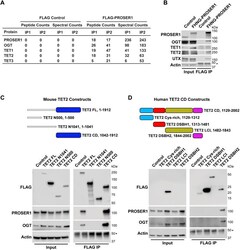
- Figure 2. PROSER1 interacts with members of the TET family of DNA demethylases and OGT. (A) FLAG-PROSER1 IP followed by mass spectrometry identifies the glycosyltransferase OGT and all three TET family proteins. (B) Western blot of FLAG IP from FLAG-HA-NeonGreen-PROSER1 ( FHNG-PROSER1 ) knock-in HEK293 cells confirming interaction of PROSER1 with OGT, TET1, TET2, and UTX. Wild-type (WT) HEK293 cells were used as an IP control. Nuclear extracts were used as input. Actin was used as a loading control for the inputs. The asterisk indicates the IgG heavy chain. (C) Top: Domain structure and constructs of mouse TET2. Blue: cysteine-rich dioxygenase (CD) domain. TET2 FL, full length construct. TET2 N500, N-terminal 500 amino acids (aa). TET2 N1041, N-terminal 1,041 aa. TET2 CD, TET2 cysteine-rich dioxygenase domain. Bottom: HEK293 cells were transiently transfected with a series of FLAG-tagged mouse TET2 constructs as shown on the top. WB of FLAG IPs from total cell lysates showing interaction of PROSER1 and OGT with TET2 FL and TET2 CD. Total cell lysates were used as inputs. Actin was used as a loading control for the inputs. The asterisk indicates the IgG heavy chain. (D) Top: Domain structure and constructs of the human TET2 cysteine-rich dioxygenase (CD) domain. Light blue, cysteine-rich domain. Red, double-stranded beta-helix (DSBH1) domain 1. Olive, low-complexity insert (LCI) region. Purple, double-stranded beta-helix (DSBH2) domain 2. Bottom: HEK293 cells were transiently
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
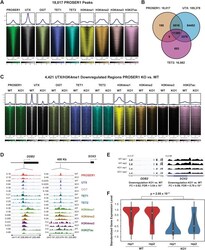
- Figure 4. PROSER1 regulates the chromatin association of TET1/2 to mediate UTX/H3K4me1-dependent enhancer activity. (A) Heatmaps centered on 18,017 reproducible PROSER1 binding sites obtained from three different PROSER1 antibodies in WT HEK293 cells. Occupancy of PROSER1, UTX, OGT, TET1, TET2, H3K4me1, H3K4me2, H3K4me3, and H3K27ac is displayed. (B) Venn diagram depicting the overlap between reproducible PROSER1, UTX, and TET2 peaks in WT HEK293 cells. (C) Heatmaps centered on 4,421 UTX/H3K4me1-down-regulated regions in PROSER1 KO compared with WT cells. Occupancy of PROSER1, UTX, OGT, TET1, TET2, H3K4me1, H3K4me2, H3K4me3, and H3K27ac is displayed. (D) Genome browser tracks depicting the ChIP-seq profiles of PROSER1, UTX, OGT, TET1, TET2, H3K4me1, H3K4me2, H3K4me3, and H3K27ac in WT and PROSER1 KO HEK293 cells at the DDB2 promoter (left) and SOX2 enhancer (right). (E) Genome browser tracks showing the RNA-seq profiles of the DDB2 and SOX2 genes in WT and PROSER1 KO HEK293 cells. Two replicates are displayed for each genotype. (F) Violin plots showing transcriptional down-regulation of the 400 genes associated with the 4,421 regions displaying lower enrichment of UTX and H3K4me1 in PROSER1 KO compared with WT cells. Expression values were standardized for each gene across the samples. The median interquartile range and the 1.5x interquartile range are shown for each violin plot.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry