Antibody data
- Antibody Data
- Antigen structure
- References [20]
- Comments [0]
- Validations
- Immunocytochemistry [5]
- Immunohistochemistry [2]
- Flow cytometry [6]
- Other assay [6]
Submit
Validation data
Reference
Comment
Report error
- Product number
- MA3-813 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- CRABP1 Monoclonal Antibody (C-1)
- Antibody type
- Monoclonal
- Antigen
- Other
- Description
- MA3-813 detects cellular retinoic acid binding protein I (CRABPI) from human, mouse, monkey, feline, bovine, rabbit, rat, turtle and chicken tissues. This antibody does not react with chameleon, goldfish and frog. This antibody does not cross-react with cellular retinol binding protein (CRBP), performic acid oxidized CRBP, interphotoreceptor retinoid binding protein or retinol binding protein. MA3-813 has been successfully used in Western blot, immunohistochemistry and ELISA procedures. By Western blot, this antibody detects a 16 kDa protein representing CRABPI from rat retinal supernatant. Immunohistochemical staining of CRABPI in rat retina with MA3-813 results in its localization to amacrine somata and laminae in the inner plexiform layer. The MA3-813 antigen is oxidized CRABPI from bovine retina.
- Reactivity
- Human, Mouse, Rat, Bovine, Chicken/Avian, Feline, Rabbit
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- C-1
- Vial size
- 100 μL
- Concentration
- 1 mg/mL
- Storage
- -20°C, Avoid Freeze/Thaw Cycles
Submitted references Myofibroblast transcriptome indicates SFRP2(hi) fibroblast progenitors in systemic sclerosis skin.
Proteome analysis of the effects of all-trans retinoic acid on human germ cell tumor cell lines.
Isolation and properties of an in vitro human outer blood-retinal barrier model.
Retinoic acid orchestrates fibroblast growth factor signalling to drive embryonic stem cell differentiation.
LIF removal increases CRABPI and CRABPII transcripts in embryonic stem cells cultured in retinol or 4-oxoretinol.
Overexpression of CRABPI in suprabasal keratinocytes enhances the proliferation of epidermal basal keratinocytes in mouse skin topically treated with all-trans retinoic acid.
Distribution of the cellular retinoic acid binding protein CRABP-I in the developing chick optic tectum.
Establishment of a human in vitro model of the outer blood-retinal barrier.
In cerebrospinal fluid ER chaperones ERp57 and calreticulin bind beta-amyloid.
Differential gene expression in ovarian carcinoma: identification of potential biomarkers.
SFRP1 modulates retina cell differentiation through a beta-catenin-independent mechanism.
Sources and sink of retinoic acid in the embryonic chick retina: distribution of aldehyde dehydrogenase activities, CRABP-I, and sites of retinoic acid inactivation.
Retinoic acid promotes differentiation of photoreceptors in vitro.
Retinoic acid promotes differentiation of photoreceptors in vitro.
The developing organ of Corti contains retinoic acid and forms supernumerary hair cells in response to exogenous retinoic acid in culture.
The developing organ of Corti contains retinoic acid and forms supernumerary hair cells in response to exogenous retinoic acid in culture.
Localization of cellular retinoic acid-binding protein to amacrine cells of rat retina.
Immunolocalization of cellular retinoic acid binding protein to Müller cells and/or a subpopulation of GABA-positive amacrine cells in retinas of different species.
Immunolocalization of cellular retinol-, retinaldehyde- and retinoic acid-binding proteins in rat retina during pre- and postnatal development.
Immunolocalization of cellular retinol-, retinaldehyde- and retinoic acid-binding proteins in rat retina during pre- and postnatal development.
Tabib T, Huang M, Morse N, Papazoglou A, Behera R, Jia M, Bulik M, Monier DE, Benos PV, Chen W, Domsic R, Lafyatis R
Nature communications 2021 Jul 19;12(1):4384
Nature communications 2021 Jul 19;12(1):4384
Proteome analysis of the effects of all-trans retinoic acid on human germ cell tumor cell lines.
Honecker F, Rohlfing T, Harder S, Braig M, Gillis AJ, Glaesener S, Barett C, Bokemeyer C, Buck F, Brümmendorf TH, Looijenga LH, Balabanov S
Journal of proteomics 2014 Jan 16;96:300-13
Journal of proteomics 2014 Jan 16;96:300-13
Isolation and properties of an in vitro human outer blood-retinal barrier model.
Hamilton RD, Leach L
Methods in molecular biology (Clifton, N.J.) 2011;686:401-16
Methods in molecular biology (Clifton, N.J.) 2011;686:401-16
Retinoic acid orchestrates fibroblast growth factor signalling to drive embryonic stem cell differentiation.
Stavridis MP, Collins BJ, Storey KG
Development (Cambridge, England) 2010 Mar;137(6):881-90
Development (Cambridge, England) 2010 Mar;137(6):881-90
LIF removal increases CRABPI and CRABPII transcripts in embryonic stem cells cultured in retinol or 4-oxoretinol.
Lane MA, Xu J, Wilen EW, Sylvester R, Derguini F, Gudas LJ
Molecular and cellular endocrinology 2008 Jan 2;280(1-2):63-74
Molecular and cellular endocrinology 2008 Jan 2;280(1-2):63-74
Overexpression of CRABPI in suprabasal keratinocytes enhances the proliferation of epidermal basal keratinocytes in mouse skin topically treated with all-trans retinoic acid.
Tang XH, Vivero M, Gudas LJ
Experimental cell research 2008 Jan 1;314(1):38-51
Experimental cell research 2008 Jan 1;314(1):38-51
Distribution of the cellular retinoic acid binding protein CRABP-I in the developing chick optic tectum.
Propping C, Mönig B, Luksch H, Mey J
Brain research 2007 Sep 7;1168:21-31
Brain research 2007 Sep 7;1168:21-31
Establishment of a human in vitro model of the outer blood-retinal barrier.
Hamilton RD, Foss AJ, Leach L
Journal of anatomy 2007 Dec;211(6):707-16
Journal of anatomy 2007 Dec;211(6):707-16
In cerebrospinal fluid ER chaperones ERp57 and calreticulin bind beta-amyloid.
Erickson RR, Dunning LM, Olson DA, Cohen SJ, Davis AT, Wood WG, Kratzke RA, Holtzman JL
Biochemical and biophysical research communications 2005 Jun 24;332(1):50-7
Biochemical and biophysical research communications 2005 Jun 24;332(1):50-7
Differential gene expression in ovarian carcinoma: identification of potential biomarkers.
Hibbs K, Skubitz KM, Pambuccian SE, Casey RC, Burleson KM, Oegema TR Jr, Thiele JJ, Grindle SM, Bliss RL, Skubitz AP
The American journal of pathology 2004 Aug;165(2):397-414
The American journal of pathology 2004 Aug;165(2):397-414
SFRP1 modulates retina cell differentiation through a beta-catenin-independent mechanism.
Esteve P, Trousse F, Rodríguez J, Bovolenta P
Journal of cell science 2003 Jun 15;116(Pt 12):2471-81
Journal of cell science 2003 Jun 15;116(Pt 12):2471-81
Sources and sink of retinoic acid in the embryonic chick retina: distribution of aldehyde dehydrogenase activities, CRABP-I, and sites of retinoic acid inactivation.
Mey J, McCaffery P, Klemeit M
Brain research. Developmental brain research 2001 Apr 30;127(2):135-48
Brain research. Developmental brain research 2001 Apr 30;127(2):135-48
Retinoic acid promotes differentiation of photoreceptors in vitro.
Kelley MW, Turner JK, Reh TA
Development (Cambridge, England) 1994 Aug;120(8):2091-102
Development (Cambridge, England) 1994 Aug;120(8):2091-102
Retinoic acid promotes differentiation of photoreceptors in vitro.
Kelley MW, Turner JK, Reh TA
Development (Cambridge, England) 1994 Aug;120(8):2091-102
Development (Cambridge, England) 1994 Aug;120(8):2091-102
The developing organ of Corti contains retinoic acid and forms supernumerary hair cells in response to exogenous retinoic acid in culture.
Kelley MW, Xu XM, Wagner MA, Warchol ME, Corwin JT
Development (Cambridge, England) 1993 Dec;119(4):1041-53
Development (Cambridge, England) 1993 Dec;119(4):1041-53
The developing organ of Corti contains retinoic acid and forms supernumerary hair cells in response to exogenous retinoic acid in culture.
Kelley MW, Xu XM, Wagner MA, Warchol ME, Corwin JT
Development (Cambridge, England) 1993 Dec;119(4):1041-53
Development (Cambridge, England) 1993 Dec;119(4):1041-53
Localization of cellular retinoic acid-binding protein to amacrine cells of rat retina.
Gaur VP, de Leeuw AM, Milam AH, Saari JC
Experimental eye research 1990 May;50(5):505-11
Experimental eye research 1990 May;50(5):505-11
Immunolocalization of cellular retinoic acid binding protein to Müller cells and/or a subpopulation of GABA-positive amacrine cells in retinas of different species.
Milam AH, De Leeuw AM, Gaur VP, Saari JC
The Journal of comparative neurology 1990 Jun 1;296(1):123-9
The Journal of comparative neurology 1990 Jun 1;296(1):123-9
Immunolocalization of cellular retinol-, retinaldehyde- and retinoic acid-binding proteins in rat retina during pre- and postnatal development.
De Leeuw AM, Gaur VP, Saari JC, Milam AH
Journal of neurocytology 1990 Apr;19(2):253-64
Journal of neurocytology 1990 Apr;19(2):253-64
Immunolocalization of cellular retinol-, retinaldehyde- and retinoic acid-binding proteins in rat retina during pre- and postnatal development.
De Leeuw AM, Gaur VP, Saari JC, Milam AH
Journal of neurocytology 1990 Apr;19(2):253-64
Journal of neurocytology 1990 Apr;19(2):253-64
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
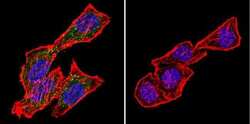
- Experimental details
- Immunofluorescent analysis of CRABPI using Anti-CRABPI Monoclonal Antibody (C-1) (Product # MA3-813) shows staining in Hela Cells. CRABPI staining (green), F-Actin staining with Phalloidin (red) and nuclei with DAPI (blue) is shown. Cells were grown on chamber slides and fixed with formaldehyde prior to staining. Cells were probed without (control) or with or an antibody recognizing CRABPI (Product # MA3-813) at a dilution of 1:200 over night at 4°C, washed with PBS and incubated with a DyLight-488 conjugated secondary antibody (Product # 35503, Goat Anti-Mouse). Images were taken at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
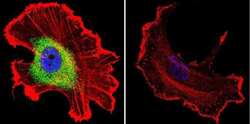
- Experimental details
- Immunofluorescent analysis of CRABPI using Anti-CRABPI Monoclonal Antibody (C-1) (Product # MA3-813) shows staining in MCF-7 Cells. CRABPI staining (green), F-Actin staining with Phalloidin (red) and nuclei with DAPI (blue) is shown. Cells were grown on chamber slides and fixed with formaldehyde prior to staining. Cells were probed without (control) or with or an antibody recognizing CRABPI (Product # MA3-813) at a dilution of 1:100 over night at 4°C, washed with PBS and incubated with a DyLight-488 conjugated secondary antibody (Product # 35503, Goat Anti-Mouse). Images were taken at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
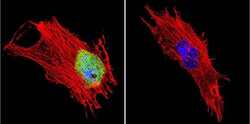
- Experimental details
- Immunofluorescent analysis of CRABPI using Anti-CRABPI Monoclonal Antibody (C-1) (Product # MA3-813) shows staining in NIH-3T3 Cells. CRABPI staining (green), F-Actin staining with Phalloidin (red) and nuclei with DAPI (blue) is shown. Cells were grown on chamber slides and fixed with formaldehyde prior to staining. Cells were probed without (control) or with or an antibody recognizing CRABPI (Product # MA3-813) at a dilution of 1:200 over night at 4°C, washed with PBS and incubated with a DyLight-488 conjugated secondary antibody (Product # 35503, Goat Anti-Mouse). Images were taken at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
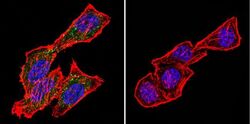
- Experimental details
- Immunofluorescent analysis of CRABPI using Anti-CRABPI Monoclonal Antibody (C-1) (Product # MA3-813) shows staining in Hela Cells. CRABPI staining (green), F-Actin staining with Phalloidin (red) and nuclei with DAPI (blue) is shown. Cells were grown on chamber slides and fixed with formaldehyde prior to staining. Cells were probed without (control) or with or an antibody recognizing CRABPI (Product # MA3-813) at a dilution of 1:200 over night at 4°C, washed with PBS and incubated with a DyLight-488 conjugated secondary antibody (Product # 35503, Goat Anti-Mouse). Images were taken at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
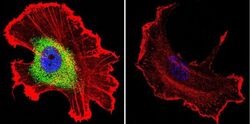
- Experimental details
- Immunofluorescent analysis of CRABPI using Anti-CRABPI Monoclonal Antibody (C-1) (Product # MA3-813) shows staining in MCF-7 Cells. CRABPI staining (green), F-Actin staining with Phalloidin (red) and nuclei with DAPI (blue) is shown. Cells were grown on chamber slides and fixed with formaldehyde prior to staining. Cells were probed without (control) or with or an antibody recognizing CRABPI (Product # MA3-813) at a dilution of 1:100 over night at 4°C, washed with PBS and incubated with a DyLight-488 conjugated secondary antibody (Product # 35503, Goat Anti-Mouse). Images were taken at 60X magnification.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
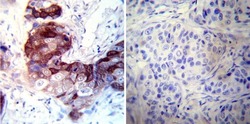
- Experimental details
- Immunohistochemistry was performed on cancer biopsies of deparaffinized Human breast carcinoma tissues. To expose target proteins, heat induced antigen retrieval was performed using 10mM sodium citrate (pH6.0) buffer, microwaved for 8-15 minutes. Following antigen retrieval tissues were blocked in 3% BSA-PBS for 30 minutes at room temperature. Tissues were then probed at a dilution of 1:200 with a mouse monoclonal antibody recognizing CRABPI (Product # MA3-813) or without primary antibody (negative control) overnight at 4°C in a humidified chamber. Tissues were washed extensively with PBST and endogenous peroxidase activity was quenched with a peroxidase suppressor. Detection was performed using a biotin-conjugated secondary antibody and SA-HRP, followed by colorimetric detection using DAB. Tissues were counterstained with hematoxylin and prepped for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
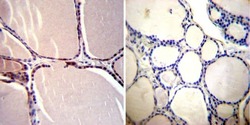
- Experimental details
- Immunohistochemistry was performed on normal deparaffinized Human thyroid tissue tissues. To expose target proteins, heat induced antigen retrieval was performed using 10mM sodium citrate (pH6.0) buffer, microwaved for 8-15 minutes. Following antigen retrieval tissues were blocked in 3% BSA-PBS for 30 minutes at room temperature. Tissues were then probed at a dilution of 1:200 with a mouse monoclonal antibody recognizing CRABPI (Product # MA3-813) or without primary antibody (negative control) overnight at 4°C in a humidified chamber. Tissues were washed extensively with PBST and endogenous peroxidase activity was quenched with a peroxidase suppressor. Detection was performed using a biotin-conjugated secondary antibody and SA-HRP, followed by colorimetric detection using DAB. Tissues were counterstained with hematoxylin and prepped for mounting.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
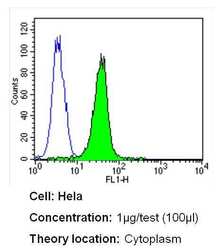
- Experimental details
- Flow cytometry analysis of CRABPI in Hela cells (green) compared to an isotype control (blue). Cells were harvested, adjusted to a concentration of 1-5x10^6 cells/mL, fixed with 2% paraformaldehyde and washed with PBS. Cells were blocked with a 2% solution of BSA-PBS for 30 min at room temperature and incubated with a CRABPI monoclonal antibody (Product # MA3-813) at a dilution of 1 µg/test for 40 min at room temperature. Cells were then incubated for 40 min at room temperature in the dark using a Dylight 488-conjugated secondary antibody and re-suspended in PBS for FACS analysis.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
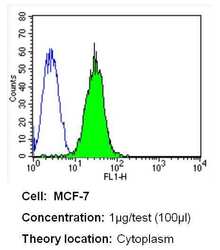
- Experimental details
- Flow cytometry analysis of CRABPI in MCF-7 cells (green) compared to an isotype control (blue). Cells were harvested, adjusted to a concentration of 1-5x10^6 cells/mL, fixed with 2% paraformaldehyde and washed with PBS. Cells were blocked with a 2% solution of BSA-PBS for 30 min at room temperature and incubated with a CRABPI monoclonal antibody (Product # MA3-813) at a dilution of 1 µg/test for 40 min at room temperature. Cells were then incubated for 40 min at room temperature in the dark using a Dylight 488-conjugated secondary antibody and re-suspended in PBS for FACS analysis.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
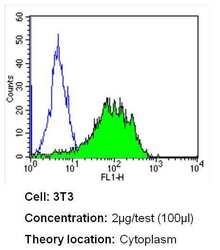
- Experimental details
- Flow cytometry analysis of CRABPI in NIH-3T3 cells (green) compared to an isotype control (blue). Cells were harvested, adjusted to a concentration of 1-5x10^6 cells/mL, fixed with 2% paraformaldehyde and washed with PBS. Cells were blocked with a 2% solution of BSA-PBS for 30 min at room temperature and incubated with a CRABPI monoclonal antibody (Product # MA3-813) at a dilution of 2 µg/test for 40 min at room temperature. Cells were then incubated for 40 min at room temperature in the dark using a Dylight 488-conjugated secondary antibody and re-suspended in PBS for FACS analysis.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
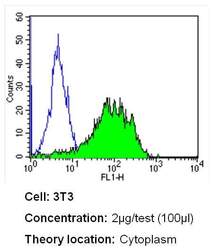
- Experimental details
- Flow cytometry analysis of CRABPI in NIH-3T3 cells (green) compared to an isotype control (blue). Cells were harvested, adjusted to a concentration of 1-5x10^6 cells/mL, fixed with 2% paraformaldehyde and washed with PBS. Cells were blocked with a 2% solution of BSA-PBS for 30 min at room temperature and incubated with a CRABPI monoclonal antibody (Product # MA3-813) at a dilution of 2 µg/test for 40 min at room temperature. Cells were then incubated for 40 min at room temperature in the dark using a Dylight 488-conjugated secondary antibody and re-suspended in PBS for FACS analysis.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
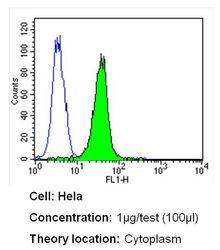
- Experimental details
- Flow cytometry analysis of CRABPI in Hela cells (green) compared to an isotype control (blue). Cells were harvested, adjusted to a concentration of 1-5x10^6 cells/mL, fixed with 2% paraformaldehyde and washed with PBS. Cells were blocked with a 2% solution of BSA-PBS for 30 min at room temperature and incubated with a CRABPI monoclonal antibody (Product # MA3-813) at a dilution of 1 µg/test for 40 min at room temperature. Cells were then incubated for 40 min at room temperature in the dark using a Dylight 488-conjugated secondary antibody and re-suspended in PBS for FACS analysis.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
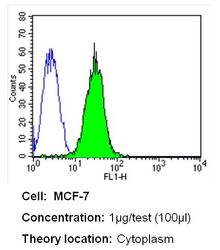
- Experimental details
- Flow cytometry analysis of CRABPI in MCF-7 cells (green) compared to an isotype control (blue). Cells were harvested, adjusted to a concentration of 1-5x10^6 cells/mL, fixed with 2% paraformaldehyde and washed with PBS. Cells were blocked with a 2% solution of BSA-PBS for 30 min at room temperature and incubated with a CRABPI monoclonal antibody (Product # MA3-813) at a dilution of 1 µg/test for 40 min at room temperature. Cells were then incubated for 40 min at room temperature in the dark using a Dylight 488-conjugated secondary antibody and re-suspended in PBS for FACS analysis.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
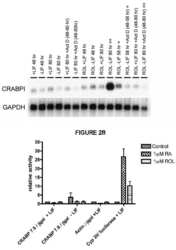
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
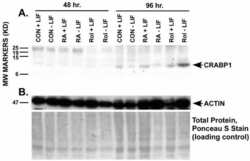
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
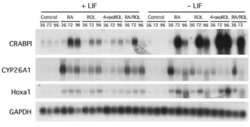
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry