Antibody data
- Antibody Data
- Antigen structure
- References [7]
- Comments [0]
- Validations
- Immunocytochemistry [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA021319 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA021319, RRID:AB_1856554
- Product name
- Anti-SAMD9
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human SAMD9, Gene description: sterile alpha motif domain containing 9, Alternative Gene Names: C7orf5, FLJ20073, KIAA2004, Validated applications: WB, IHC, ICC, Uniprot ID: Q5K651, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.5 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references Myxoma virus lacking the host range determinant M062 stimulates cGAS-dependent type 1 interferon response and unique transcriptomic changes in human monocytes/macrophages
Non-replicating Vaccinia Virus TianTan Strain (NTV) Translation Arrest of Viral Late Protein Synthesis Associated With Anti-viral Host Factor SAMD9.
RNA granules associated with SAMD9-mediated poxvirus restriction are similar to antiviral granules in composition but do not require TIA1 for poxvirus restriction
An interaction domain in human SAMD9 is essential for myxoma virus host-range determinant M062 antagonism of host anti-viral function
Somatic mutations and progressive monosomy modify SAMD9-related phenotypes in humans
SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7
miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9
Bartee E, Conrad S, Raza T, Peterson E, Liem J, Connor R, Nounamo B, Cannon M, Liu J
PLOS Pathogens 2022;18(9):e1010316
PLOS Pathogens 2022;18(9):e1010316
Non-replicating Vaccinia Virus TianTan Strain (NTV) Translation Arrest of Viral Late Protein Synthesis Associated With Anti-viral Host Factor SAMD9.
Zhao Y, Zhao L, Huang P, Ren J, Zhang P, Tian H, Tan W
Frontiers in cellular and infection microbiology 2020;10:116
Frontiers in cellular and infection microbiology 2020;10:116
RNA granules associated with SAMD9-mediated poxvirus restriction are similar to antiviral granules in composition but do not require TIA1 for poxvirus restriction
Meng X, Xiang Y
Virology 2019;529
Virology 2019;529
An interaction domain in human SAMD9 is essential for myxoma virus host-range determinant M062 antagonism of host anti-viral function
Nounamo B, Li Y, O’Byrne P, Kearney A, Khan A, Liu J
Virology 2017;503
Virology 2017;503
Somatic mutations and progressive monosomy modify SAMD9-related phenotypes in humans
Buonocore F, Kühnen P, Suntharalingham J, Del Valle I, Digweed M, Stachelscheid H, Khajavi N, Didi M, Brady A, Blankenstein O, Procter A, Dimitri P, Wales J, Ghirri P, Knöbl D, Strahm B, Erlacher M, Wlodarski M, Chen W, Kokai G, Anderson G, Morrogh D, Moulding D, McKee S, Niemeyer C, Grüters A, Achermann J
Journal of Clinical Investigation 2017;127(5):1700-1713
Journal of Clinical Investigation 2017;127(5):1700-1713
SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7
Narumi S, Amano N, Ishii T, Katsumata N, Muroya K, Adachi M, Toyoshima K, Tanaka Y, Fukuzawa R, Miyako K, Kinjo S, Ohga S, Ihara K, Inoue H, Kinjo T, Hara T, Kohno M, Yamada S, Urano H, Kitagawa Y, Tsugawa K, Higa A, Miyawaki M, Okutani T, Kizaki Z, Hamada H, Kihara M, Shiga K, Yamaguchi T, Kenmochi M, Kitajima H, Fukami M, Shimizu A, Kudoh J, Shibata S, Okano H, Miyake N, Matsumoto N, Hasegawa T
Nature Genetics 2016;48(7):792-797
Nature Genetics 2016;48(7):792-797
miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9
WU L, PU X, WANG Q, CAO J, XU F, XU L, LI K
Oncology Letters 2016;11(2):945-952
Oncology Letters 2016;11(2):945-952
No comments: Submit comment
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
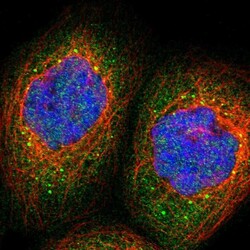
- Experimental details
- Immunofluorescent staining of human cell line A-431 shows localization to cytosol & vesicles.
- Sample type
- Human
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry