Antibody data
- Antibody Data
- Antigen structure
- References [15]
- Comments [0]
- Validations
- Immunocytochemistry [2]
- Immunohistochemistry [1]
- Flow cytometry [5]
- Other assay [11]
Submit
Validation data
Reference
Comment
Report error
- Product number
- MA5-13548 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- CD81 Monoclonal Antibody (1.3.3.22)
- Antibody type
- Monoclonal
- Antigen
- Other
- Description
- MA5-13548 targets CD81 in WB, FACS, FN, ICC/IF, and IHC (F) applications and shows reactivity with Human and Rat samples. The MA5-13548 immunogen is b cell line derived from a Burkitt lymphoma.
- Reactivity
- Human, Rat
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- 1.3.3.22
- Vial size
- 500 μL
- Concentration
- 0.2 mg/mL
- Storage
- 4°C
Submitted references Characterization of plasma exosomal microRNAs in responding to radiotherapy of human esophageal squamous cell carcinoma.
Functional Characterization of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells.
Anti-Human CD9 Fab Fragment Antibody Blocks the Extracellular Vesicle-Mediated Increase in Malignancy of Colon Cancer Cells.
Exosomes in serum‑free cultures of THP‑1 macrophages infected with Mycobacterium tuberculosis.
Human Mesenchymal Stromal Cell-Derived Exosomes Promote In Vitro Wound Healing by Modulating the Biological Properties of Skin Keratinocytes and Fibroblasts and Stimulating Angiogenesis.
Electro-Acupuncture Promotes the Differentiation of Endogenous Neural Stem Cells via Exosomal microRNA 146b After Ischemic Stroke.
Plasma Small Extracellular Vesicles Derived miR-21-5p and miR-92a-3p as Potential Biomarkers for Hepatocellular Carcinoma Screening.
Optimized method for extraction of exosomes from human primary muscle cells.
Inhalation of lung spheroid cell secretome and exosomes promotes lung repair in pulmonary fibrosis.
Integrated Dual-Mode Chromatography to Enrich Extracellular Vesicles from Plasma.
Serum milk fat globule-EGF factor 8 (MFG-E8) as a diagnostic and prognostic biomarker in patients with hepatocellular carcinoma.
JC Polyomavirus Uses Extracellular Vesicles To Infect Target Cells.
Human embryonic stem cells extracellular vesicles and their effects on immortalized human retinal Müller cells.
Inflammasome Proteins in Serum and Serum-Derived Extracellular Vesicles as Biomarkers of Stroke.
Exosomal microRNAs derived from colorectal cancer-associated fibroblasts: role in driving cancer progression.
Miao N, Cai W, Ding S, Liu Y, Chen W, Sun T
Molecular medicine reports 2022 Sep;26(3)
Molecular medicine reports 2022 Sep;26(3)
Functional Characterization of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells.
Fan X, Cyganek L, Nitschke K, Uhlig S, Nuhn P, Bieback K, Duerschmied D, El-Battrawy I, Zhou X, Akin I
International journal of molecular sciences 2022 Jul 31;23(15)
International journal of molecular sciences 2022 Jul 31;23(15)
Anti-Human CD9 Fab Fragment Antibody Blocks the Extracellular Vesicle-Mediated Increase in Malignancy of Colon Cancer Cells.
Santos MF, Rappa G, Fontana S, Karbanová J, Aalam F, Tai D, Li Z, Pucci M, Alessandro R, Morimoto C, Corbeil D, Lorico A
Cells 2022 Aug 10;11(16)
Cells 2022 Aug 10;11(16)
Exosomes in serum‑free cultures of THP‑1 macrophages infected with Mycobacterium tuberculosis.
Biadglegne F, Rademacher P, De Sulbaran YGJ, König B, Rodloff AC, Zedler U, Dorhoi A, Sack U
Molecular medicine reports 2021 Nov;24(5)
Molecular medicine reports 2021 Nov;24(5)
Human Mesenchymal Stromal Cell-Derived Exosomes Promote In Vitro Wound Healing by Modulating the Biological Properties of Skin Keratinocytes and Fibroblasts and Stimulating Angiogenesis.
Tutuianu R, Rosca AM, Iacomi DM, Simionescu M, Titorencu I
International journal of molecular sciences 2021 Jun 9;22(12)
International journal of molecular sciences 2021 Jun 9;22(12)
Electro-Acupuncture Promotes the Differentiation of Endogenous Neural Stem Cells via Exosomal microRNA 146b After Ischemic Stroke.
Zhang S, Jin T, Wang L, Liu W, Zhang Y, Zheng Y, Lin Y, Yang M, He X, Lin H, Chen L, Tao J
Frontiers in cellular neuroscience 2020;14:223
Frontiers in cellular neuroscience 2020;14:223
Plasma Small Extracellular Vesicles Derived miR-21-5p and miR-92a-3p as Potential Biomarkers for Hepatocellular Carcinoma Screening.
Sorop A, Iacob R, Iacob S, Constantinescu D, Chitoiu L, Fertig TE, Dinischiotu A, Chivu-Economescu M, Bacalbasa N, Savu L, Gheorghe L, Dima S, Popescu I
Frontiers in genetics 2020;11:712
Frontiers in genetics 2020;11:712
Optimized method for extraction of exosomes from human primary muscle cells.
Le Gall L, Ouandaogo ZG, Anakor E, Connolly O, Butler Browne G, Laine J, Duddy W, Duguez S
Skeletal muscle 2020 Jul 8;10(1):20
Skeletal muscle 2020 Jul 8;10(1):20
Inhalation of lung spheroid cell secretome and exosomes promotes lung repair in pulmonary fibrosis.
Dinh PC, Paudel D, Brochu H, Popowski KD, Gracieux MC, Cores J, Huang K, Hensley MT, Harrell E, Vandergriff AC, George AK, Barrio RT, Hu S, Allen TA, Blackburn K, Caranasos TG, Peng X, Schnabel LV, Adler KB, Lobo LJ, Goshe MB, Cheng K
Nature communications 2020 Feb 28;11(1):1064
Nature communications 2020 Feb 28;11(1):1064
Integrated Dual-Mode Chromatography to Enrich Extracellular Vesicles from Plasma.
Van Deun J, Jo A, Li H, Lin HY, Weissleder R, Im H, Lee H
Advanced biosystems 2020 Dec;4(12):e1900310
Advanced biosystems 2020 Dec;4(12):e1900310
Serum milk fat globule-EGF factor 8 (MFG-E8) as a diagnostic and prognostic biomarker in patients with hepatocellular carcinoma.
Shimagaki T, Yoshio S, Kawai H, Sakamoto Y, Doi H, Matsuda M, Mori T, Osawa Y, Fukai M, Yoshida T, Ma Y, Akita T, Tanaka J, Taketomi A, Hanayama R, Yoshizumi T, Mori M, Kanto T
Scientific reports 2019 Oct 31;9(1):15788
Scientific reports 2019 Oct 31;9(1):15788
JC Polyomavirus Uses Extracellular Vesicles To Infect Target Cells.
Morris-Love J, Gee GV, O'Hara BA, Assetta B, Atkinson AL, Dugan AS, Haley SA, Atwood WJ
mBio 2019 Apr 9;10(2)
mBio 2019 Apr 9;10(2)
Human embryonic stem cells extracellular vesicles and their effects on immortalized human retinal Müller cells.
Peng Y, Baulier E, Ke Y, Young A, Ahmedli NB, Schwartz SD, Farber DB
PloS one 2018;13(3):e0194004
PloS one 2018;13(3):e0194004
Inflammasome Proteins in Serum and Serum-Derived Extracellular Vesicles as Biomarkers of Stroke.
Kerr N, García-Contreras M, Abbassi S, Mejias NH, Desousa BR, Ricordi C, Dietrich WD, Keane RW, de Rivero Vaccari JP
Frontiers in molecular neuroscience 2018;11:309
Frontiers in molecular neuroscience 2018;11:309
Exosomal microRNAs derived from colorectal cancer-associated fibroblasts: role in driving cancer progression.
Bhome R, Goh RW, Bullock MD, Pillar N, Thirdborough SM, Mellone M, Mirnezami R, Galea D, Veselkov K, Gu Q, Underwood TJ, Primrose JN, De Wever O, Shomron N, Sayan AE, Mirnezami AH
Aging 2017 Dec 28;9(12):2666-2694
Aging 2017 Dec 28;9(12):2666-2694
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
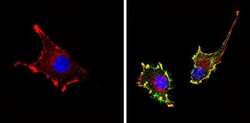
- Experimental details
- Immunofluorescent analysis of CD81 (green) showing staining in the membrane of U-87 MG cells (right) compared to a negative control without primary antibody (left). Formalin-fixed cells were permeabilized with 0.1% Triton X-100 in TBS for 5-10 minutes and blocked with 3% BSA-PBS for 30 minutes at room temperature. Cells were probed with a CD81 monoclonal antibody (Product # MA5-13548) in 3% BSA-PBS at a dilution of 1:20 and incubated overnight at 4 ºC in a humidified chamber. Cells were washed with PBST and incubated with a DyLight-conjugated secondary antibody in PBS at room temperature in the dark. F-actin (red) was stained with a fluorescent red phalloidin and nuclei (blue) were stained with Hoechst or DAPI. Images were taken at a magnification of 60x.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
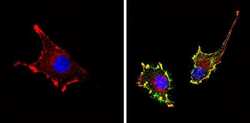
- Experimental details
- Immunofluorescent analysis of CD81 (green) showing staining in the membrane of U-87 MG cells (right) compared to a negative control without primary antibody (left). Formalin-fixed cells were permeabilized with 0.1% Triton X-100 in TBS for 5-10 minutes and blocked with 3% BSA-PBS for 30 minutes at room temperature. Cells were probed with a CD81 monoclonal antibody (Product # MA5-13548) in 3% BSA-PBS at a dilution of 1:20 and incubated overnight at 4 ºC in a humidified chamber. Cells were washed with PBST and incubated with a DyLight-conjugated secondary antibody in PBS at room temperature in the dark. F-actin (red) was stained with a fluorescent red phalloidin and nuclei (blue) were stained with Hoechst or DAPI. Images were taken at a magnification of 60x.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Frozen human tonsil stained with CD81 antibody using peroxidase-conjugate and AEC chromogen. Note cell membrane staining of lymphocytes.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
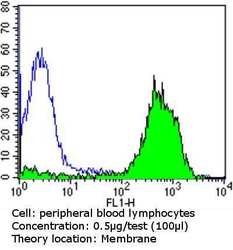
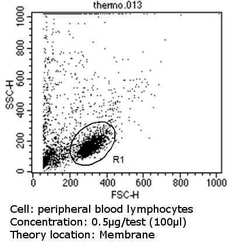
- Experimental details
- Flow cytometry analysis of CD81 in peripheral blood mononuclear cells compared to an isotype control (blue). Human blood was collected, combined with a hydrophilic polysaccharide, centrifuged, transferred to a conical tube and washed with PBS. 50 µL of cell solution was added to each tube at a dilution of 2x10^7 cells/mL, followed by the addition of 50 µL of isotype control and primary antibody (Product # MA5-13548) at a dilution of 0.5 µg/test. Cells were incubated for 30 min at 4ºC and washed with a cell buffer, followed by incubation with a DyLight 488-conjugated goat anti-mouse IgG (H+L) secondary for 30 min at 4ºC in the dark. FACS analysis was performed using 400 µL of cell buffer.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- Flow cytometry analysis of CD81 in peripheral blood mononuclear cells compared to an isotype control (blue). Human blood was collected, combined with a hydrophilic polysaccharide, centrifuged, transferred to a conical tube and washed with PBS. 50 µL of cell solution was added to each tube at a dilution of 2x10^7 cells/mL, followed by the addition of 50 µL of isotype control and primary antibody (Product # MA5-13548) at a dilution of 0.5 µg/test. Cells were incubated for 30 min at 4ºC and washed with a cell buffer, followed by incubation with a DyLight 488-conjugated goat anti-mouse IgG (H+L) secondary for 30 min at 4ºC in the dark. FACS analysis was performed using 400 µL of cell buffer.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
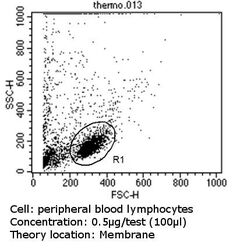
- Experimental details
- Flow cytometry analysis of CD81 in peripheral blood mononuclear cells compared to an isotype control (blue). Human blood was collected, combined with a hydrophilic polysaccharide, centrifuged, transferred to a conical tube and washed with PBS. 50 µL of cell solution was added to each tube at a dilution of 2x10^7 cells/mL, followed by the addition of 50 µL of isotype control and primary antibody (Product # MA5-13548) at a dilution of 0.5 µg/test. Cells were incubated for 30 min at 4ºC and washed with a cell buffer, followed by incubation with a DyLight 488-conjugated goat anti-mouse IgG (H+L) secondary for 30 min at 4ºC in the dark. FACS analysis was performed using 400 µL of cell buffer.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
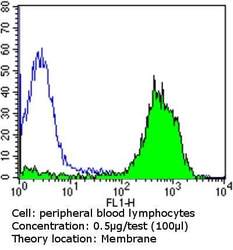
- Experimental details
- Flow cytometry analysis of CD81 in peripheral blood mononuclear cells compared to an isotype control (blue). Human blood was collected, combined with a hydrophilic polysaccharide, centrifuged, transferred to a conical tube and washed with PBS. 50 µL of cell solution was added to each tube at a dilution of 2x10^7 cells/mL, followed by the addition of 50 µL of isotype control and primary antibody (Product # MA5-13548) at a dilution of 0.5 µg/test. Cells were incubated for 30 min at 4ºC and washed with a cell buffer, followed by incubation with a DyLight 488-conjugated goat anti-mouse IgG (H+L) secondary for 30 min at 4ºC in the dark. FACS analysis was performed using 400 µL of cell buffer.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
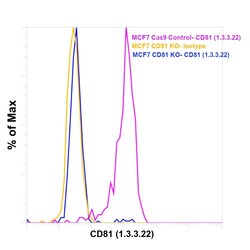
- Experimental details
- Knockout of CD81 was achieved by CRISPR-Cas9 genome editing using LentiArray™ Lentiviral sgRNA (Product # A32042, Assay ID CRISPR661098_LV) and LentiArray Cas9 Lentivirus (Product # A32064). Flow cytometry analysis of CD81 was performed by staining MCF-7 CD81 Knock out cells with 0.5 µg Mouse IgG1 kappa Isotype Control (P3.6.2.8.1), PE, eBioscience™ (Product # 12-4714-42, yellow histogram) or 0.5 µg CD81 Monoclonal Antibody (1.3.3.22) (Product # MA5-13548, blue histogram). MCF-7 Cas9 control cells were also stained with CD81 Monoclonal Antibody (1.3.3.22) (Product # MA5-13548, pink histogram). Loss of signal was observed in the CD81 KO cells stained with CD81 antibody clone 1.3.3.22 but not in the control Cas9 cells. Viable cells were used for analysis, as determined by Fixable Viability Dye eFluor™ 780 (Product # 65-0865-18). (Product # 65-0865-18).
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
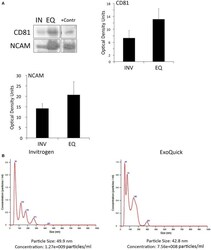
- Experimental details
- Figure 2 EV characterization in serum from stroke patients . (A) Representative immunoblot of CD81 and NCAM positive EV isolated with the Invitrogen Kit (IN) and the ExoQuick Kit (EQ). +Contr: Positive control of isolated EV. Quantification of CD81- and NCAM-positive EV isolated from serum with the Invitrogen kit (INV) and the ExoQuick kit (EQ). Nanoparticle tracking analysis of isolated serum-derived EV. (B) Nanoparticle tracking analysis predicts size distribution and concentration of particles in serum-derived EV samples isolated with the Invitrogen kit and the ExoQuick kit.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
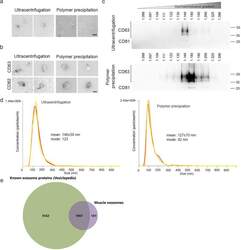
- Experimental details
- Fig. 4 Validation of exosome extraction strategy . For each experiment, exosomes were extracted from the culture medium of 100 x 10 6 myoblasts differentiated into myotubes for 3 days using either the ultracentrifugation or the polymer precipitation. The culture medium was non-supplemented DMEM (without serum). a Cup-shaped vesicles were observed by electron microscopy with both extraction protocols. bar = 100 nm. b Both extractions show vesicles that are positive for CD63 and CD82 by electron microscopy. Bar = 100 nm. c Exosome extracts were loaded on iodixanol gradients as described in material and methods. Western blot results are shown for CD63 and CD81 in twelve fractions for the iodixanol gradient. Top panel: exosomes extracted by ultracentrifugation. Bottom panel: exosomes extracted by polymer-based precipitation. Exosomes were detected at a density of 1.15-1.27 g.ml -1 . d Nanosight analyses show similar-sized vesicles using both strategies, from 100-200 nm, with a greater number of particles being detected when using the polymer extraction. e Proteomic analysis of muscle exosomes. Venn diagram showing the overlap between muscle exosomes and proteins known to be detected by mass spectrometry in exosomes (Vesiclepedia, Exocarta database [-])
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
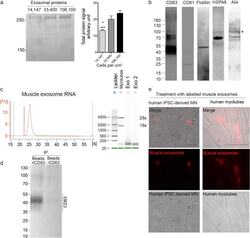
- Experimental details
- Fig. 5 Polymer-based precipitation efficiently extracts functional exosomes from 7.5 x 106 cells. a SDS-page protein quantification showing that the greatest efficiency in terms of exosomal protein per cell plated was obtained when exosomes were extracted from differentiated myoblasts at a density of 33,400 cells.cm -2 . Differentiated myoblasts were plated at 14,147 (lane 1), 33,400 (lane 2), or 106,100 (lane 3) cells per cm 2 . Right panel: representative SDS-page stained with Coomassie. Left panel: protein concentration measurements in secreted exosomes from cells plated at a different density. *, ***, P < 0.05 and P < 0.001, significantly different from 33,400 and 106,100 cells.cm 2 . ( n = 4, 3, 4 per condition). b Muscle exosomes were positive for CD63, CD81, Flotillin, HSPA8, and Alix. c mRNA was detectable with a clean profile from polymer-precipitated exosomes of 7.5 x 10 6 differentiated myoblasts. No 18 s and 28 s RNA were detected, indicating that there were no RNA contaminants from dead cells. Inset panel: Representative electrophoresis obtain with Agilent 2100 Bioanalyzer for myotubes and exosome RNA extract. d Western blot showing that polymer precipitated exosomes from 7.5 x 10 6 differentiated myoblasts can be used to pull down a specific subpopulation such as CD63 positive exosomes (+/-CD63 = with/without anti-CD63 antibody). e Polymer-precipitated exosomes (pre-stained with PKH26 following extraction; red channel) were capable of integrating into myotubes o
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
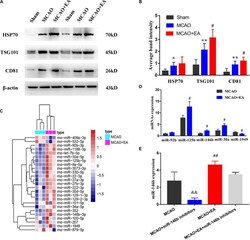
- Experimental details
- FIGURE 1 Electro-acupuncture promotes the release of exosomal miR-146b in peri-ischemic striatum. (A) The expression of exosome-associated markers HSP70, TSG101, and CD81 were analyzed by Western blotting. (B) The expression of markers were quantified on the basis of average band intensity ( n = 4). (C) MiRNA microarray analysis of exosomes from peri-ischemic striatum and clustering of 25 significantly altered miRNAs with >=1.5-fold difference in expression between MCAO and MCAO+EA groups. (D) Four miRNAs were significantly upregulated and 1 miRNA were downregulated in exosomes of the MCAO+EA group compared with the MCAO group were checked by RT-qPCR ( n = 3). (E) The expression of miR-146b in tissues of peri-ischemic striatum were analyzed form MCAO, MCAO+miR-146b inhibitors, MCAO+EA, MCAO+EA+miR-146b inhibitors groups ( n = 3). Data represent means +- SD of the results of representative experiment. * P < 0.05, MCAO vs. Sham; ** P < 0.01, MCAO vs. Sham; # P < 0.05 and ## P < 0.01, MCAO vs. MCAO+EA; && P < 0.01, MCAO vs. MCAO+miR-146b inhibitors; MCAO+EA vs. MCAO+EA+miR-146b inhibitors.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
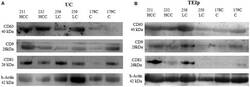
- Experimental details
- FIGURE 4 Positive detection for specific exosomal surface markers (CD 63, CD9, and CD81) and for the control-positive marker (b-actin) by immunoblotting. (A) exosomes isolated by UC and (B) TEIp kit.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
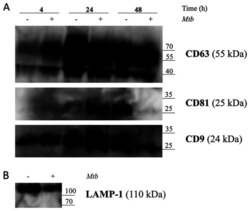
- Experimental details
- Figure 5. Release of exosomes from Mtb H37Rv-infected or naive THP-1 cells. THP-1 macrophages were infected with Mtb H37Rv (Mtb + ) for 4 h (MOI 5) in DCM-UG medium for 4, 24 and 48 h at 37degC and in the presence of 5% CO 2 . Exosomes were extracted from cell culture supernatants using total exosome isolation reagent and were further analyzed by western blot analysis for the exosomal protein markers, (A) CD63, CD81, CD9 and (B) LAMP-1, which served as a positive control for exosomes signal and Mtb proteins [Ag85 and Mpt64 and recombinant Mtb Mpt64 protein (His-tag)]. Data from one representative experiment out of at least three independent experiments are shown. MOI, multiplicity of infection; Mtb, Mycobacterium tuberculosis ; DCM-UG, ultra-centrifuged CellGenix (r) GMP DC Medium; LAMP-1, lysosomal associated membrane protein-1; Ag85, mycobacterial antigen 85.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
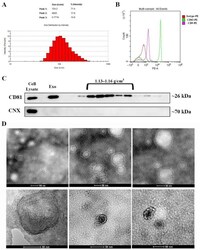
- Experimental details
- Figure 1 Characterization of exosomes isolated from bone marrow-derived MSC conditioned medium. ( A ) Histogram showing the distribution of the hydrodynamic diameter of the isolated exosomes by dynamic light scattering analysis. ( B ) Expression of exosomal markers CD63 and CD9 as revealed by flow cytometry and ( C ) CD81 as identified via Western blot for the whole pellet (Exo) as well as for the sucrose fractions derived from it. Note the absence of contaminants from the endoplasmic reticulum as indicated by the lack of calnexin (CNX) both in the exosomes (Exo) pellet and the sucrose fractions. ( D ) Transmission electron microscopy images depicting donut-shaped structures up to 100 nm (negative staining with phosphotungstic acid--upper panel and uranyl acetate--lower panel).
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
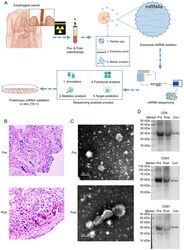
- Experimental details
- Figure 1. Exosome miRNA validation and sequencing. (A) The experimental workflow of characterization of plasma exosome miRNAs for the radiotherapy of human esophageal cancer. (B) H&E staining of esophageal squamous cell carcinoma samples (magnification, x100) from patients of pre- and post-radiotherapy. (C) Characterization of plasma exosomes. Morphological observation of exosomes by transmission electron microscopy. Scale bar=200 nm. (D) Expression levels of CD9, CD63 and CD81 were identified using western blot analysis. miRNA, microRNA.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
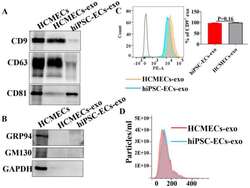
- Experimental details
- Characterization of the purified exosomes from hiPSC-EC and HCMEC conditioned media. Exosomes isolated from cell culture media of hiPSC-ECs and HCMECs. ( A ) Western blot showed the detection of exosome markers CD9, CD63, and CD81 in isolated exosomes. ( B ) Glucose-regulated protein 94 (GRP94) and Golgi marker GM130 were not found in exosomes. ( C ) Flow cytometry analysis using exosome marker CD9. ( D ) The size distribution of isolated exosomes was measured by nanoparticle tracking analysis (NTA) in hiPSC-ECs and HCMECs.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
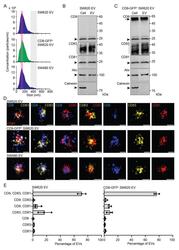
- Experimental details
- Characterization of EVs released by SW620, CD9-GFP + SW620, and SW480 cells. ( A - E ) EVs were recovered from the conditioned media of SW620, CD9-GFP + SW620, and SW480 cells by differential centrifugation, and the resulting 200,000x g pellets were analyzed by the ZetaView particle analyzer ( A ), immunoblotting ( B , C ), and dSTORM ( D , E ). The concentration and size of EVs derived from the indicated cells are shown ( A ). Note the presence of a common population of small particles ( 5000 EVs per experiment). Note that small and large EVs derived from SW480 cells were also quantified ( Supplementary Figure S2 ). Scale bars, 50 nm.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
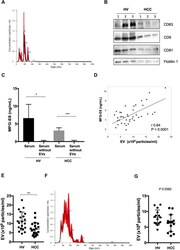
- Experimental details
- Figure 5 Serum MFG-E8 levels and EVs in HVs and HCC patients. ( A ) Extracellular vesicles (EVs) isolated using the Tim4 affinity method were analyzed by nanoparticle tracking analysis (NTA) using NanoSight LM10. ( B ) Expression of the EV markers CD63, CD9, CD81 and flotillin 1 by western blotting. Full-length blots are presented in Supplementary Fig. 2 . ( C ) Comparision of serum milk fat globule-EGF factor 8 (MFG-E8) levels before and after EV isolation by the Tim4 affinity method. ( D ) Correlation between serum MFG-E8 levels and the concentration of serum EVs isolated using the Tim4 affinity method. Sera were drawn from healthy volunteers (HVs) (n = 20) and hepatocellular carcinoma (HCC) patients (n = 20). ( E ) Comparative analysis of serum EVs from HVs and HCC patients by NTA. ( F ) EVs isolated using the ultracentrifugation method were analyzed by NTA using NanoSight LM10. ( G ) Comparative analysis of serum EVs isolated by the ultracentrifugation method from HCC patients (n = 15) and HVs (n = 15). *p < 0.05, **p < 0.01, ***p < 0.001 by Kruskal-Wallis test with Dunn's multiple comparison test.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry