Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Immunocytochemistry [3]
- Immunohistochemistry [2]
- Other assay [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-22096 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- PIGR Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant full-length protein
- Description
- Recommended positive controls: Human milk. Store product as a concentrated solution. Centrifuge briefly prior to opening the vial.
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 0.39 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references Loss of synergistic transcriptional feedback loops drives diverse B-cell cancers.
Andrews JM, Pyfrom SC, Schmidt JA, Koues OI, Kowalewski RA, Grams NR, Sun JJ, Berman LR, Duncavage EJ, Lee YS, Cashen AF, Oltz EM, Payton JE
EBioMedicine 2021 Sep;71:103559
EBioMedicine 2021 Sep;71:103559
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
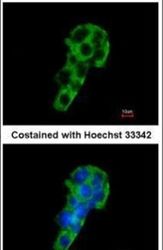
- Experimental details
- Immunofluorescent analysis of PIGR in methanol-fixed HepG2 cells using a PIGR polyclonal antibody (Product # PA5-22096) at a 1:200 dilution.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
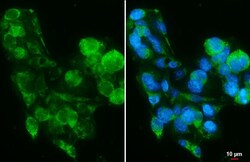
- Experimental details
- PIGR Polyclonal Antibody detects PIGR protein by immunofluorescent analysis. Sample: HepG2 cells were fixed in 4% paraformaldehyde at RT for 15 min. Green: PIGR stained by PIGR Polyclonal Antibody (Product # PA5-22096) diluted at 1:500. Blue: Fluoroshield with DAPI . Scale bar= 10 µm.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
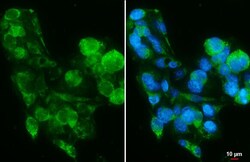
- Experimental details
- PIGR Polyclonal Antibody detects PIGR protein by immunofluorescent analysis. Sample: HepG2 cells were fixed in 4% paraformaldehyde at RT for 15 min. Green: PIGR stained by PIGR Polyclonal Antibody (Product # PA5-22096) diluted at 1:500. Blue: Fluoroshield with DAPI . Scale bar= 10 µm.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
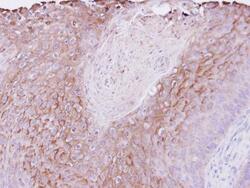
- Experimental details
- Immunohistochemical analysis of paraffin-embedded Cal27 xenograft , using PIGR (Product # PA5-22096) antibody at 1:500 dilution. Antigen Retrieval: EDTA based buffer, pH 8.0, 15 min.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
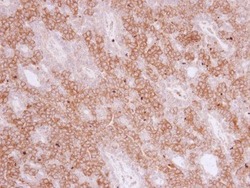
- Experimental details
- Immunohistochemistry (Paraffin) analysis of PIGR was performed in paraffin-embedded human lung adenocarcinoma tissue using PIGR Polyclonal Antibody (Product # PA5-22096) at a dilution of 1:500.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
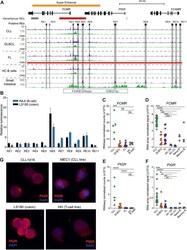
- Experimental details
- Fig. 3 A novel super-enhancer associated with dysregulated high expression of PIGR and FCMR mRNA and protein. A) Genome browser screenshot shows signal for histone modifications (ChIPseq) and chromatin accessibility (FAIREseq - WUSM samples; DNase-seq - small intestine sample from Roadmap Epigenomic Project) for the FCMR / PIGR locus. Regulatory elements that were tested in luciferase assays are highlighted in gray. GeneHancer regulatory elements (red - promoter; gray - enhancer; darker color indicates higher confidence) and the identified super-enhancer are also shown. All tracks are RPKM normalized. Regions of copy number alteration (CNA) are shown by a red bar (amplifications identified in FL samples). B) Luciferase reporter assay results for all regulatory elements shown in ( A ). Assays were read in triplicate and performed at least twice. Mean with standard deviation shown. C-F) Expression of FCMR ( C-D ) and PIGR ( E-F ) determined by RNAseq ( C&E ) and RNA microarray ( D&F ) for BCL subtypes and healthy control B cells and B cell subsets (germinal center (GC) centroblasts (CB), centrocytes (CC), naive and memory B cells). RNAseq analyzed by DESeq2; microarrays analyzed by limma. * adj p > 0.05, ** adj p < 0.01, *** adj p < 0.001, **** adj p < 0.0001, ns not significant. G) Immunofluorescence microscopy for PIGR in purified primary CLL B-cells, MEC1 (CLL cell line), LS180 (colorectal cancer cell line), and HH (T-cell lymphoma cell line). Fig 3
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry