Antibody data
- Antibody Data
- Antigen structure
- References [2]
- Comments [0]
- Validations
- Immunocytochemistry [5]
- Immunohistochemistry [7]
- Other assay [3]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA3-16829 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- NuMA Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant full-length protein
- Description
- Suggested positive control: Hela whole cell extract.
- Reactivity
- Human, Mouse, Rat
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- Conc. Not Determined
- Storage
- -20°C or -80°C if preferred
Submitted references WDR62 regulates spindle dynamics as an adaptor protein between TPX2/Aurora A and katanin.
Human Schwann cells exhibit long-term cell survival, are not tumorigenic and promote repair when transplanted into the contused spinal cord.
Huang J, Liang Z, Guan C, Hua S, Jiang K
The Journal of cell biology 2021 Aug 2;220(8)
The Journal of cell biology 2021 Aug 2;220(8)
Human Schwann cells exhibit long-term cell survival, are not tumorigenic and promote repair when transplanted into the contused spinal cord.
Bastidas J, Athauda G, De La Cruz G, Chan WM, Golshani R, Berrocal Y, Henao M, Lalwani A, Mannoji C, Assi M, Otero PA, Khan A, Marcillo AE, Norenberg M, Levi AD, Wood PM, Guest JD, Dietrich WD, Bartlett Bunge M, Pearse DD
Glia 2017 Aug;65(8):1278-1301
Glia 2017 Aug;65(8):1278-1301
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
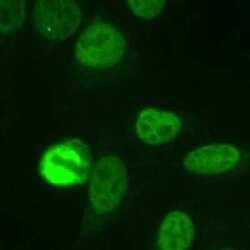
- Experimental details
- Immunocytochemistry analysis of NuMA in Human HeLa cells fixed with 3.5% formaldehyde. Samples were incubated in NuMA polyclonal antibody (Product # PA3-16829) using a dilution of 1:1000. The staining pattern shows the typical pattern for NuMA in the cell nucleus during interphase and on spindles during mitosis.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
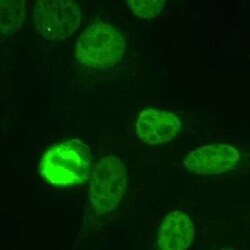
- Experimental details
- Immunocytochemistry analysis of NuMA in Human HeLa cells fixed with 3.5% formaldehyde. Samples were incubated in NuMA polyclonal antibody (Product # PA3-16829) using a dilution of 1:1000. The staining pattern shows the typical pattern for NuMA in the cell nucleus during interphase and on spindles during mitosis.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
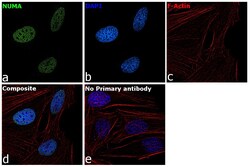
- Experimental details
- Immunofluorescence analysis of NuMA was performed using HeLa cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 15 minutes, and blocked with 2% BSA for 1 hour at room temperature. The cells were labeled with NuMA Polyclonal Antibody (Product # PA3-16829) at 1:200 dilution in 0.1% BSA, incubated at 4 degree celsius overnight and then labeled with Goat anti-Rabbit IgG (H+L) Superclonal™ Recombinant Secondary Antibody, Alexa Fluor® 488 conjugate (Product # A27034) at a dilution of 1:2000 for 45 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with ProLong™ Diamond Antifade Mountant with DAPI (Product # P36962). F-actin (Panel c: red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image showing nuclear localization. Panel e represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
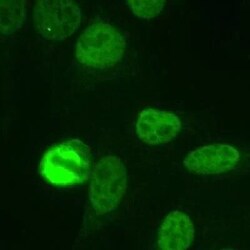
- Experimental details
- Immunocytochemistry analysis of NuMA in Human HeLa cells fixed with 3.5% formaldehyde. Samples were incubated in NuMA polyclonal antibody (Product # PA3-16829) using a dilution of 1:1000. The staining pattern shows the typical pattern for NuMA in the cell nucleus during interphase and on spindles during mitosis.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
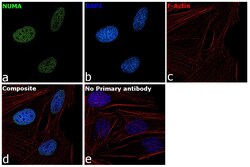
- Experimental details
- Immunofluorescence analysis of NuMA was performed using HeLa cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 15 minutes, and blocked with 2% BSA for 1 hour at room temperature. The cells were labeled with NuMA Polyclonal Antibody (Product # PA3-16829) at 1:200 dilution in 0.1% BSA, incubated at 4 degree celsius overnight and then labeled with Goat anti-Rabbit IgG (Heavy Chain) Superclonal™ Recombinant Secondary Antibody, Alexa Fluor® 488 conjugate (Product # A27034) at a dilution of 1:2000 for 45 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with ProLong™ Diamond Antifade Mountant with DAPI (Product # P36962). F-actin (Panel c: red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image showing nuclear localization. Panel e represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Immunohistochemical analysis of NuMA in formalin-fixed paraffin-embedded tissue section of human normal brain. Samples were incubated in NuMA polyclonal antibody (Product # PA3-16829) using a dilution of 5 µg/mL. The cells in the brain tissue depicted strong specific nuclear along with weak cytoplasmic immuno-positivity for NuMA protein. 10X Magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
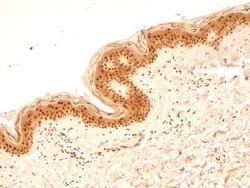
- Experimental details
- Immunohistochemical analysis of NuMA in formalin-fixed paraffin-embedded tissue section of human normal skin. Samples were incubated in NuMA polyclonal antibody (Product # PA3-16829) using a dilution of 5 µg/mL. Almost all the cells in the epidermal layer and the cells in the connective tissue of dermal layer depicted very strong nuclear along with moderate cytoplasmic immuno-positivity for NuMA protein. 10X Magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
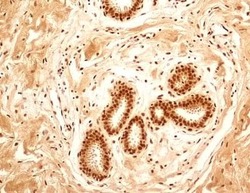
- Experimental details
- Immunohistochemical analysis of NuMA in formalin-fixed paraffin-embedded tissue section of human normal breast. Samples were incubated in NuMA polyclonal antibody (Product # PA3-16829) using a dilution of 5 µg/mL. The breast ductal/acinar epithelial cells, the myoepithelial cells and some cells in the surrounding connective tissue depicted a strong nuclear along with moderate cytoplasmic immuno-positivity for NuMA protein. The intra-lobular and the surrounding connective tissue also developed weak/potentially non-specific staining. 10X Magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Immunohistochemical analysis of NuMA in formalin-fixed paraffin-embedded tissue section of human normal spleen. Samples were incubated in NuMA polyclonal antibody (Product # PA3-16829) using a dilution of 5 µg/mL. Most of the cells developed a strong nuclear along with moderate cytoplasmic immuno-positivity for NuMA protein. 10X Magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
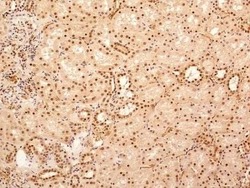
- Experimental details
- Immunohistochemical analysis of NuMA in formalin-fixed paraffin-embedded tissue section of human normal kidney. Samples were incubated in NuMA polyclonal antibody (Product # PA3-16829) using a dilution of 5 µg/mL. The cells of various tubules/ducts and Bowmans capsule in the renal tissue depicted strong nuclear along with relatively weak cytoplasmic immuno-positivity for NuMA protein. 10X Magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
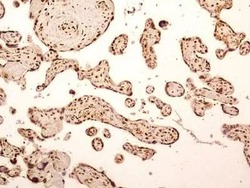
- Experimental details
- Immunohistochemical analysis of NuMA in formalin-fixed paraffin-embedded tissue section of human placenta. Samples were incubated in NuMA polyclonal antibody (Product # PA3-16829) using a dilution of 5 µg/mL. Strong nuclear with moderate cytoplasmic immuno-reactivity of NuMA was observed in syncytiotrophoblast, cytiotrophoblasts, Hofbauer cells and the endothelial cells of chorionic villi capillaries. 10X Magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
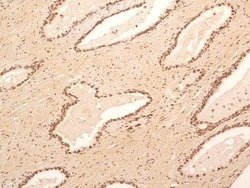
- Experimental details
- Immunohistochemical analysis of NuMA in formalin-fixed paraffin-embedded tissue section of human normal prostate. Samples were incubated in NuMA polyclonal antibody (Product # PA3-16829) using a dilution of 5 µg/mL. The prostatic alveolar glandular epithelium as well as the surrounding fibromuscular stroma cells showed a strong nuclear immuno-reactivity for NuMA protein. Almost all the cells developed some cytoplasmic staining also. 10X Magnification.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
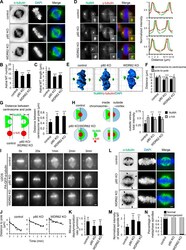
- Experimental details
- Figure 2. WDR62-katanin regulates spindle organization and dynamics. (A) Immunofluorescence staining for alpha-tubulin and DAPI in control and indicated knockout (KO) HeLa cells. Maximum intensity projections of Z series with 30 stacks at 0.11-um steps are shown. (B and C) Quantification of astral MT number and length as shown in A. n = 28, 24, and 20 spindle poles for control, p80 knockout, and WDR62 knockout, respectively. (D) Left: Immunofluorescence staining for NuMA, gamma-tubulin, and DAPI in control and the indicated knockout HeLa cells. Insets show enlargements of the boxed areas. Right: Normalized line-scan intensity profiles of NuMA (green) and gamma-tubulin (red) corresponding to the white dashed lines in left images. (E) 3D reconstruction of images shown in D. The length of the grid in the images is 3.65 um. (F) Quantification of centrosome-to-centrosome distance and pole-to-pole distance (see Materials and methods for details) in control and indicated knockout HeLa cells. n = 40 cells for all conditions. (G) Left: Schematic representation of the distance between centrosome and pole (denoted by double-headed arrow), as measured from the center of gamma-tubulin (gamma-tub) spot (red) to the distal edge of NuMA spot (green). Right: Quantification of the distance between centrosome and pole in control and indicated knockout HeLa cells. n = 80 poles for all conditions. (H) Left: Schematic representation of outside and inside. The centers of two semicircular ROIs were
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
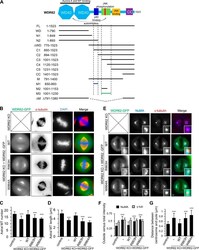
- Figure S2. Overview of WDR62 deletion mutants used in this study and the rescue of spindle defects by WT WDR62-GFP but not its katanin-binding deficient mutants. (A) Schematic illustration of the functional domains of WDR62 and summary of all the deletion mutants used in this study. WD40 domain (WD; light blue): MT binding and Aurora A binding; M1 (dark blue): p80 binding; M3: (green): autoinhibitory domain that interacts with WD40 domain. The horizontal lines were drawn to indicate the region spanned by the corresponding fragments (M1: dark blue; M3: green; others: black). The vertical dashed lines were drawn to indicate the boundaries of fragments M1 and M3. Note that JNK phosphorylation site T1053 (red) was located within the autoinhibitory M3 fragment. M, middle region; CC, coiled-coil. (B) Immunofluorescence staining of alpha-tubulin and DAPI in WDR62 knockout (KO) HeLa cells or the knockout cells transiently transfected with GFP-tagged WT WDR62 or its indicated mutants. Maximum intensity projections of Z series with 30 stacks at 0.11-um steps are shown. (C and D) Quantification of the number and length of astral MTs shown in B. From left to right, n = 20, 20, 22, 20, and 20 spindle poles. (E) Immunofluorescence staining of NuMA and gamma-tubulin in WDR62 knockout HeLa cells or the knockout cells transiently transfected with GFP-tagged WT WDR62 or its indicated mutants. Insets show enlargement of boxed areas. (F and G) Quantification of ""outside"" versus total intensity
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry Immunoprecipitation
Immunoprecipitation