51-5100
antibody from Invitrogen Antibodies
Targeting: HDAC2
KDAC2, RPD3, YAF1
 Western blot
Western blot ELISA
ELISA Immunocytochemistry
Immunocytochemistry Immunoprecipitation
Immunoprecipitation Immunohistochemistry
Immunohistochemistry Flow cytometry
Flow cytometry Chromatin Immunoprecipitation
Chromatin Immunoprecipitation Other assay
Other assayAntibody data
- Antibody Data
- Antigen structure
- References [26]
- Comments [0]
- Validations
- Immunocytochemistry [4]
- Immunohistochemistry [3]
- Flow cytometry [1]
- Other assay [18]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 51-5100 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- HDAC2 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Synthetic peptide
- Reactivity
- Human, Mouse
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μg
- Concentration
- 0.25 mg/mL
- Storage
- -20°C
Submitted references HDAC1/2 Control Proliferation and Survival in Adult Epidermis and Pre‒Basal Cell Carcinoma through p16 and p53.
Combined genomic and proteomic approaches reveal DNA binding sites and interaction partners of TBX2 in the developing lung.
Cancer Stem-Cell Marker CD44v9-Positive Cells Arise From Helicobacter pylori-Infected CAPZA1-Overexpressing Cells.
PP2A negatively regulates the hypertrophic response by dephosphorylating HDAC2 S394 in the heart.
Transient expression of ZBTB32 in anti-viral CD8+ T cells limits the magnitude of the effector response and the generation of memory.
Inhibition of class I histone deacetylase activity represses matrix metalloproteinase-2 and -9 expression and preserves LV function postmyocardial infarction.
Contrasting roles for MyoD in organizing myogenic promoter structures during embryonic skeletal muscle development.
Spatial re-organization of myogenic regulatory sequences temporally controls gene expression.
Ctbp2 Modulates NuRD-Mediated Deacetylation of H3K27 and Facilitates PRC2-Mediated H3K27me3 in Active Embryonic Stem Cell Genes During Exit from Pluripotency.
Deacetylase inhibitors repress STAT5-mediated transcription by interfering with bromodomain and extra-terminal (BET) protein function.
Redox-sensitive gene-regulatory events controlling aberrant matrix metalloproteinase-1 expression.
Transcriptional repression of ER through hMAPK dependent histone deacetylation by class I HDACs.
Epigenetic modifications induced by Blimp-1 Regulate CD8⁺ T cell memory progression during acute virus infection.
pVHL-mediated transcriptional repression of c-Myc by recruitment of histone deacetylases.
Histone modifiers, YY1 and p300, regulate the expression of cartilage-specific gene, chondromodulin-I, in mesenchymal stem cells.
Histone deacetylases facilitate sodium/calcium exchanger up-regulation in adult cardiomyocytes.
Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1.
The hTERT and hTERC telomerase gene promoters are activated by the second exon of the adenoviral protein, E1A, identifying the transcriptional corepressor CtBP as a potential repressor of both genes.
Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis.
Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis.
Heregulin regulates the ability of the ErbB3-binding protein Ebp1 to bind E2F promoter elements and repress E2F-mediated transcription.
Histone deacetylase inhibitors activate INK4d gene through Sp1 site in its promoter.
Class I histone deacetylase-selective novel synthetic inhibitors potently inhibit human tumor proliferation.
The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2.
Repression of E2F1-mediated transcription by the ErbB3 binding protein Ebp1 involves histone deacetylases.
Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells.
Zhu X, Leboeuf M, Liu F, Grachtchouk M, Seykora JT, Morrisey EE, Dlugosz AA, Millar SE
The Journal of investigative dermatology 2022 Jan;142(1):77-87.e10
The Journal of investigative dermatology 2022 Jan;142(1):77-87.e10
Combined genomic and proteomic approaches reveal DNA binding sites and interaction partners of TBX2 in the developing lung.
Lüdtke TH, Wojahn I, Kleppa MJ, Schierstaedt J, Christoffels VM, Künzler P, Kispert A
Respiratory research 2021 Mar 17;22(1):85
Respiratory research 2021 Mar 17;22(1):85
Cancer Stem-Cell Marker CD44v9-Positive Cells Arise From Helicobacter pylori-Infected CAPZA1-Overexpressing Cells.
Tsugawa H, Kato C, Mori H, Matsuzaki J, Kameyama K, Saya H, Hatakeyama M, Suematsu M, Suzuki H
Cellular and molecular gastroenterology and hepatology 2019;8(3):319-334
Cellular and molecular gastroenterology and hepatology 2019;8(3):319-334
PP2A negatively regulates the hypertrophic response by dephosphorylating HDAC2 S394 in the heart.
Yoon S, Kook T, Min HK, Kwon DH, Cho YK, Kim M, Shin S, Joung H, Jeong SH, Lee S, Kang G, Park Y, Kim YS, Ahn Y, McMullen JR, Gergs U, Neumann J, Kim KK, Kim J, Nam KI, Kim YK, Kook H, Eom GH
Experimental & molecular medicine 2018 Jul 26;50(7):1-14
Experimental & molecular medicine 2018 Jul 26;50(7):1-14
Transient expression of ZBTB32 in anti-viral CD8+ T cells limits the magnitude of the effector response and the generation of memory.
Shin HM, Kapoor VN, Kim G, Li P, Kim HR, Suresh M, Kaech SM, Wherry EJ, Selin LK, Leonard WJ, Welsh RM, Berg LJ
PLoS pathogens 2017 Aug;13(8):e1006544
PLoS pathogens 2017 Aug;13(8):e1006544
Inhibition of class I histone deacetylase activity represses matrix metalloproteinase-2 and -9 expression and preserves LV function postmyocardial infarction.
Mani SK, Kern CB, Kimbrough D, Addy B, Kasiganesan H, Rivers WT, Patel RK, Chou JC, Spinale FG, Mukherjee R, Menick DR
American journal of physiology. Heart and circulatory physiology 2015 Jun 1;308(11):H1391-401
American journal of physiology. Heart and circulatory physiology 2015 Jun 1;308(11):H1391-401
Contrasting roles for MyoD in organizing myogenic promoter structures during embryonic skeletal muscle development.
Cho OH, Mallappa C, Hernández-Hernández JM, Rivera-Pérez JA, Imbalzano AN
Developmental dynamics : an official publication of the American Association of Anatomists 2015 Jan;244(1):43-55
Developmental dynamics : an official publication of the American Association of Anatomists 2015 Jan;244(1):43-55
Spatial re-organization of myogenic regulatory sequences temporally controls gene expression.
Harada A, Mallappa C, Okada S, Butler JT, Baker SP, Lawrence JB, Ohkawa Y, Imbalzano AN
Nucleic acids research 2015 Feb 27;43(4):2008-21
Nucleic acids research 2015 Feb 27;43(4):2008-21
Ctbp2 Modulates NuRD-Mediated Deacetylation of H3K27 and Facilitates PRC2-Mediated H3K27me3 in Active Embryonic Stem Cell Genes During Exit from Pluripotency.
Kim TW, Kang BH, Jang H, Kwak S, Shin J, Kim H, Lee SE, Lee SM, Lee JH, Kim JH, Kim SY, Cho EJ, Kim JH, Park KS, Che JH, Han DW, Kang MJ, Yi EC, Youn HD
Stem cells (Dayton, Ohio) 2015 Aug;33(8):2442-55
Stem cells (Dayton, Ohio) 2015 Aug;33(8):2442-55
Deacetylase inhibitors repress STAT5-mediated transcription by interfering with bromodomain and extra-terminal (BET) protein function.
Pinz S, Unser S, Buob D, Fischer P, Jobst B, Rascle A
Nucleic acids research 2015 Apr 20;43(7):3524-45
Nucleic acids research 2015 Apr 20;43(7):3524-45
Redox-sensitive gene-regulatory events controlling aberrant matrix metalloproteinase-1 expression.
Bartling TR, Subbaram S, Clark RR, Chandrasekaran A, Kar S, Melendez JA
Free radical biology & medicine 2014 Sep;74:99-107
Free radical biology & medicine 2014 Sep;74:99-107
Transcriptional repression of ER through hMAPK dependent histone deacetylation by class I HDACs.
Plotkin A, Volmar CH, Wahlestedt C, Ayad N, El-Ashry D
Breast cancer research and treatment 2014 Sep;147(2):249-63
Breast cancer research and treatment 2014 Sep;147(2):249-63
Epigenetic modifications induced by Blimp-1 Regulate CD8⁺ T cell memory progression during acute virus infection.
Shin HM, Kapoor V, Guan T, Kaech SM, Welsh RM, Berg LJ
Immunity 2013 Oct 17;39(4):661-75
Immunity 2013 Oct 17;39(4):661-75
pVHL-mediated transcriptional repression of c-Myc by recruitment of histone deacetylases.
Hwang IY, Roe JS, Seol JH, Kim HR, Cho EJ, Youn HD
Molecules and cells 2012 Feb;33(2):195-201
Molecules and cells 2012 Feb;33(2):195-201
Histone modifiers, YY1 and p300, regulate the expression of cartilage-specific gene, chondromodulin-I, in mesenchymal stem cells.
Aoyama T, Okamoto T, Fukiage K, Otsuka S, Furu M, Ito K, Jin Y, Ueda M, Nagayama S, Nakayama T, Nakamura T, Toguchida J
The Journal of biological chemistry 2010 Sep 24;285(39):29842-50
The Journal of biological chemistry 2010 Sep 24;285(39):29842-50
Histone deacetylases facilitate sodium/calcium exchanger up-regulation in adult cardiomyocytes.
Chandrasekaran S, Peterson RE, Mani SK, Addy B, Buchholz AL, Xu L, Thiyagarajan T, Kasiganesan H, Kern CB, Menick DR
FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2009 Nov;23(11):3851-64
FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2009 Nov;23(11):3851-64
Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1.
Ohkawa Y, Marfella CG, Imbalzano AN
The EMBO journal 2006 Feb 8;25(3):490-501
The EMBO journal 2006 Feb 8;25(3):490-501
The hTERT and hTERC telomerase gene promoters are activated by the second exon of the adenoviral protein, E1A, identifying the transcriptional corepressor CtBP as a potential repressor of both genes.
Glasspool RM, Burns S, Hoare SF, Svensson C, Keith WN
Neoplasia (New York, N.Y.) 2005 Jun;7(6):614-22
Neoplasia (New York, N.Y.) 2005 Jun;7(6):614-22
Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis.
Zhu P, Martin E, Mengwasser J, Schlag P, Janssen KP, Göttlicher M
Cancer cell 2004 May;5(5):455-63
Cancer cell 2004 May;5(5):455-63
Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis.
Zhu P, Martin E, Mengwasser J, Schlag P, Janssen KP, Göttlicher M
Cancer cell 2004 May;5(5):455-63
Cancer cell 2004 May;5(5):455-63
Heregulin regulates the ability of the ErbB3-binding protein Ebp1 to bind E2F promoter elements and repress E2F-mediated transcription.
Zhang Y, Hamburger AW
The Journal of biological chemistry 2004 Jun 18;279(25):26126-33
The Journal of biological chemistry 2004 Jun 18;279(25):26126-33
Histone deacetylase inhibitors activate INK4d gene through Sp1 site in its promoter.
Yokota T, Matsuzaki Y, Miyazawa K, Zindy F, Roussel MF, Sakai T
Oncogene 2004 Jul 8;23(31):5340-9
Oncogene 2004 Jul 8;23(31):5340-9
Class I histone deacetylase-selective novel synthetic inhibitors potently inhibit human tumor proliferation.
Park JH, Jung Y, Kim TY, Kim SG, Jong HS, Lee JW, Kim DK, Lee JS, Kim NK, Kim TY, Bang YJ
Clinical cancer research : an official journal of the American Association for Cancer Research 2004 Aug 1;10(15):5271-81
Clinical cancer research : an official journal of the American Association for Cancer Research 2004 Aug 1;10(15):5271-81
The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2.
Krämer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, Brill B, Groner B, Bach I, Heinzel T, Göttlicher M
The EMBO journal 2003 Jul 1;22(13):3411-20
The EMBO journal 2003 Jul 1;22(13):3411-20
Repression of E2F1-mediated transcription by the ErbB3 binding protein Ebp1 involves histone deacetylases.
Zhang Y, Woodford N, Xia X, Hamburger AW
Nucleic acids research 2003 Apr 15;31(8):2168-77
Nucleic acids research 2003 Apr 15;31(8):2168-77
Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells.
Won J, Yim J, Kim TK
The Journal of biological chemistry 2002 Oct 11;277(41):38230-8
The Journal of biological chemistry 2002 Oct 11;277(41):38230-8
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
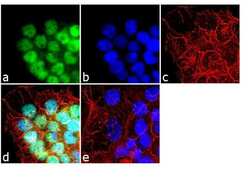
- Experimental details
- Immunofluorescence analysis of HDAC2 was done on 70% confluent log phase A431 cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 10 minutes, and blocked with 1% BSA for 1 hour at room temperature. The cells were labeled with HDAC2 Rabbit Polyclonal Antibody (Product # 51-5100) at 2 µg/mL in 0.1% BSA and incubated for 3 hours at room temperature and then labeled with Goat anti-Rabbit IgG (H+L) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate (Product # A27034) at a dilution of 1:2000 for 45 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (Product # S36938). F-actin (Panel c: red) was stained with Alexa Fluor® 555 Rhodamine Phalloidin (Product # R415, 1:300). Panel d is a merged image showing nuclear localization. Panel e is a no primary antibody control. The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
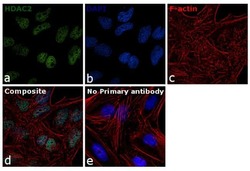
- Experimental details
- Immunofluorescence analysis of HDAC2 was performed using 70% confluent log phase HeLa cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 15 minutes, and blocked with 1% BSA for 1 hour at room temperature. The cells were labeled with HDAC2 Polyclonal Antibody (Product # 51-5100) at 5 µg/mL in 0.1% BSA, incubated at 4 degree Celsius overnight and then labeled with Goat anti-Rabbit IgG (H+L) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate (Product # A27034) at a dilution of 1:2000 for 45 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with ProLong™ Diamond Antifade Mountant with DAPI (Product # P36962). F-actin (Panel c: red) was stained with Rhodamine Phalloidin (Product # R415). Panel d represents the merged image showing nuclear localization. Panel e represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
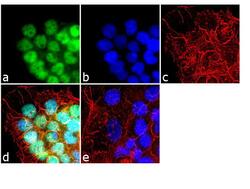
- Experimental details
- Immunofluorescence analysis of HDAC2 was done on 70% confluent log phase A431 cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 10 minutes, and blocked with 1% BSA for 1 hour at room temperature. The cells were labeled with HDAC2 Rabbit Polyclonal Antibody (Product # 51-5100) at 2 µg/mL in 0.1% BSA and incubated for 3 hours at room temperature and then labeled with Goat anti-Rabbit IgG (Heavy Chain) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate (Product # A27034) at a dilution of 1:2000 for 45 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (Product # S36938). F-actin (Panel c: red) was stained with Alexa Fluor® 555 Rhodamine Phalloidin (Product # R415, 1:300). Panel d is a merged image showing nuclear localization. Panel e is a no primary antibody control. The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
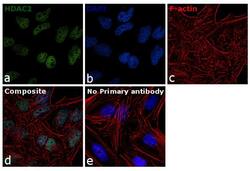
- Experimental details
- Immunofluorescence analysis of HDAC2 was performed using 70% confluent log phase HeLa cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 15 minutes, and blocked with 1% BSA for 1 hour at room temperature. The cells were labeled with HDAC2 Polyclonal Antibody (Product # 51-5100) at 5 µg/mL in 0.1% BSA, incubated at 4 degree Celsius overnight and then labeled with Goat anti-Rabbit IgG (Heavy Chain) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate (Product # A27034) at a dilution of 1:2000 for 45 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with ProLong™ Diamond Antifade Mountant with DAPI (Product # P36962). F-actin (Panel c: red) was stained with Rhodamine Phalloidin (Product # R415). Panel d represents the merged image showing nuclear localization. Panel e represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
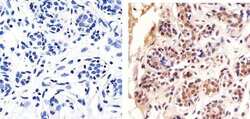
- Experimental details
- Immunohistochemistry analysis of HDAC2 showing staining in the cytoplasm and nucleus of paraffin-embedded human breast tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a Anti- HDAC2 Polyclonal Antibody (Product # 51-5100) diluted in 3% BSA-PBS at a dilution of 1:20 overnight at 4ºC in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
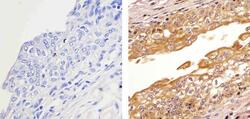
- Experimental details
- Immunohistochemistry analysis of HDAC2 showing staining in the cytoplasm and nucleus of paraffin-embedded human bladder tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a Anti- HDAC2 Polyclonal Antibody (Product # 51-5100) diluted in 3% BSA-PBS at a dilution of 1:20 overnight at 4ºC in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
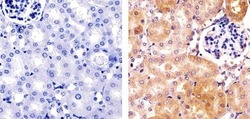
- Experimental details
- Immunohistochemistry analysis of HDAC2 showing staining in the cytoplasm and nucleus of paraffin-embedded mouse kidney tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a Anti- HDAC2 Polyclonal Antibody (Product # 51-5100) diluted in 3% BSA-PBS at a dilution of 1:100 overnight at 4ºC in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
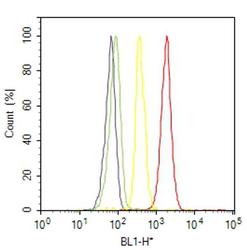
- Experimental details
- Flow cytometry analysis of HDAC2 was done on A-431 cells. Cells were fixed with 70% ethanol for 10 minutes, permeabilized with 0.25% Triton™ X-100 for 20 minutes, and blocked with 5% BSA for 30 minutes at room temperature. Cells were labeled with HDAC2 Rabbit Polyclonal Antibody (515100, red histogram) or with rabbit isotype control (yellow histogram) at 3-5 ug/million cells in 2.5% BSA. After incubation at room temperature for 2 hours, the cells were labeled with Alexa Fluor® 488 Goat Anti-Rabbit Secondary Antibody (A11008) at a dilution of 1:400 for 30 minutes at room temperature. The representative 10,000 cells were acquired and analyzed for each sample using an Attune® Acoustic Focusing Cytometer. The purple histogram represents unstained control cells and the green histogram represents no-primary-antibody control..
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
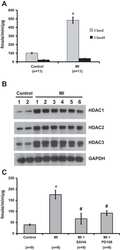
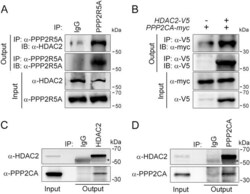
- Fig. 1 HDAC2 physically interacts with PP2A. a To confirm a physical interaction between PPP2R5A, the regulatory subunit of PP2A, and HDAC2 in NRVCs, primary cultured cardiomyocytes, 2 mg of protein was immunoprecipitated with an anti-PPP2R5A antibody, and the HDAC2 interaction was visualized with an anti-HDAC2 antibody. b-d The catalytic subunit of PP2A, PPP2CA, also bound to HDAC2. To check the interaction in H9c2 cells, a cardiomyoblast cell line, HDAC2-V5 and PPP2CA-myc were transiently transfected, and cells were lysed with 1% NP-40 lysis buffer. Immunoprecipitation was performed with anti-V5 antibody followed by immunoblotting with anti-Myc antibody. c To check the endogenous interaction in NRVCs, primary cultured cardiomyocytes were lysed in 1% NP-40 lysis buffer. Immunoprecipitation was performed with anti-HDAC2 antibody followed by immunoblotting with anti-PPP2CA antibody. d PPP2CA physically interacted with HDAC2 in mouse heart. Two milligrams of heart lysates were immunoprecipitated with anti-PPP2CA. HDAC2 successfully precipitated with PPP2CA
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
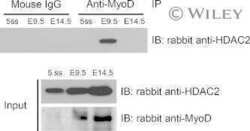
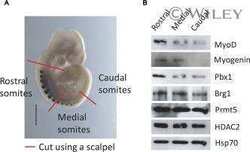
- Figure 6. MyoD and HDAC2 repress myogenic late genes at early times of differentiation. ( A ) Time course of gene expression demonstrating that bypassing MyoD by differentiating B22 cells with myogenin/Mef2D1b resulted in premature late gene activation. mRNA levels were normalized to EF1alpha levels. ( B ) ChIP time course indicating that HDAC2 did not bind to late gene regulatory sequences in myogenin/Mef2D1b differentiated B22 cells where MyoD is not expressed. The binding in the myogenin/Mef2D expressing cells at time 0 was normalized to 1. Data are presented as the average of three or more independent experiments +- standard deviation.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
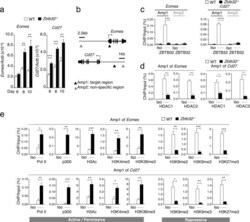
- Experimental details
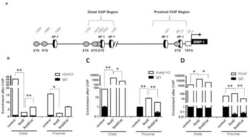
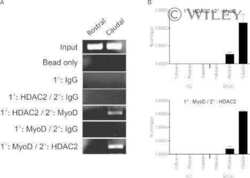
- Fig 6 ZBTB32 represses Eomes and Cd27 gene expression in CD8 + T cells by recruiting histone deacetylases 1 and 2. WT or Zbtb32 -/- P14 splenocytes were transferred into recipients, which were then infected with LCMV-Armstrong. At days 6, 8 and 10 post-infection, P14 cells were isolated and pooled from three mice per genotype for RNA isolation; chromatin was prepared at day 7 post-infection. (a) Eomes and Cd27 mRNA levels were examined by quantitative RT-PCR relative to Actb mRNA. (b) Schematic of Eomes and Cd27 gene loci showing position of specific (Amplicon 1; Amp1) and non-specific (Amplicon 2; Amp2) primers. In each case, Amp1 corresponds to putative ZBTB32 binding site. (c) The enrichment of ZBTB32 on Eomes and Cd27 genes by chromatin immunoprecipitation (ChIP). (d) The enrichment of HDAC1 and HDAC2 on Eomes and Cd27 genes by ChIP. (e) ChIP for Pol II, p300 or modified histone H3 at the Eomes and Cd27 loci. Data are a compilation of three independent experiments; error bars represent the SEM. Iso; isotype control antibody.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
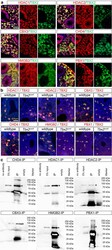
- Experimental details
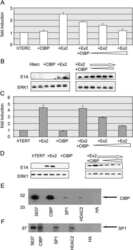
- Fig. 4 Interaction candidates are coexpressed with TBX2 in the pulmonary mesenchyme and interact in HEK293 cells. a Co-immunofluorescence analysis of candidate interaction partners (red) and TBX2 (green) on frontal sections of the right lung of E14.5 Tbx2 cre/ + embryos. Antigens are color-coded and nuclei were counterstained with DAPI (blue). Insets or selected regions in overview images are magnified in rows 2,4 and 6. b In situ proximity ligation assay of TBX2 and candidate interaction partners on 10 um frontal sections of E14.5 wildtype and Tbx2 cre/fl mutant lungs. Direct interaction is visualized by small red fluorescent dots. Larger more diffuse orange stains are due to auto-fluorescence of blood cells. Nuclei are counterstained with DAPI (blue). c Western blot analysis of co-immunoprecipitation experiments for verification of TBX2 interaction with candidate proteins on 10% SDS polyacrylamide gels. Detection was performed with an anti-TBX2 primary antibody and developed with chemoluminescence-IHC. Arrows indicate TBX2 bands. Lanes were loaded as follows: No antibody: IP without specific antibody resembling negative IP-control; 5% input: 5% of crude cell extract before precipitation; empty: no protein loaded; IP: co-immunoprecipitate with antibody for specific candidate. Expected molecular weight for TBX2.HA approx. 76.2 kDa
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
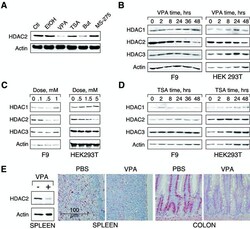
- Experimental details
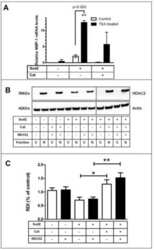
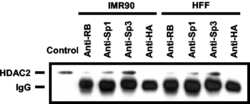
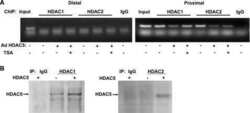
- VPA but not TSA leads to reduction of HDAC2 protein levels. ( A ) K562 human erythroleukemia cells were treated for 24 h as indicated with the HDAC inhibitors VPA (1.5 mM), TSA (100 nM), butyrate (1.5 mM) or MS-27-275 (5 muM). Amounts of HDAC2 protein were determined by western blot analysis of whole-cell extracts. Actin protein levels were determined to verify equal loading of samples. ( B ) F9 mouse teratocarcinoma or HEK293T human embryonic kidney carcinoma cells were exposed to 1 mM VPA for the indicated periods of time. Protein levels of HDAC2 as well as HDAC1, HDAC3 and actin were determined by western blot analysis. ( C ) The dose-dependent reduction of HDAC2 protein levels was determined in F9 or HEK293T cells after exposure to VPA for 30 or 24 h, respectively. ( D ) Time course analyses in F9 and HEK293T cells confirmed that TSA (100 nM) does not affect the amount of HDAC2 protein. ( E ) Reduction of HDAC2 protein levels after treatment of mice with VPA was tested by western blot analysis of tissue extracts and immunohistochemistry. Similar results were obtained in at least two sets of independent experiments.
 Explore
Explore Validate
Validate Learn
Learn