Antibody data
- Antibody Data
- Antigen structure
- References [24]
- Comments [0]
- Validations
- Western blot [1]
- Immunocytochemistry [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 18980-1-AP - Provider product page

- Provider
- Proteintech Group
- Proper citation
- Proteintech Cat#18980-1-AP, RRID:AB_10598327
- Product name
- ZCCHC11 antibody
- Antibody type
- Polyclonal
- Description
- KD/KO validated ZCCHC11 antibody (Cat. #18980-1-AP) is a rabbit polyclonal antibody that shows reactivity with human, mouse and has been validated for the following applications: IF, IP, WB,ELISA.
- Reactivity
- Human, Mouse
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 20ul, 150ul
Submitted references LINE-1 mRNA 3' end dynamics shape its biology and retrotransposition potential.
TENT2, TUT4, and TUT7 selectively regulate miRNA sequence and abundance.
Knockouts of TUT7 and 3'hExo show that they cooperate in histone mRNA maintenance and degradation.
RNA uridyl transferases TUT4/7 differentially regulate miRNA variants depending on the cancer cell type.
Extracellular microRNA 3' end modification across diverse body fluids.
Terminal uridyltransferase 7 regulates TLR4-triggered inflammation by controlling Regnase-1 mRNA uridylation and degradation.
YB1 regulates miR-205/200b-ZEB1 axis by inhibiting microRNA maturation in hepatocellular carcinoma.
Identification of RNA-binding proteins that partner with Lin28a to regulate Dnmt3a expression.
MEF2C shapes the microtranscriptome during differentiation of skeletal muscles.
Critical Roles of Translation Initiation and RNA Uridylation in Endogenous Retroviral Expression and Neural Differentiation in Pluripotent Stem Cells.
AGO-bound mature miRNAs are oligouridylated by TUTs and subsequently degraded by DIS3L2.
A Mechanism for microRNA Arm Switching Regulated by Uridylation.
A programmed wave of uridylation-primed mRNA degradation is essential for meiotic progression and mammalian spermatogenesis.
Exonuclease requirements for mammalian ribosomal RNA biogenesis and surveillance.
3' Uridylation Confers miRNAs with Non-canonical Target Repertoires.
Uridylation by TUT4/7 Restricts Retrotransposition of Human LINE-1s.
MiRAR-miRNA Activity Reporter for Living Cells.
3' Uridylation controls mature microRNA turnover during CD4 T-cell activation.
mRNA 3' uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome.
Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs.
Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4).
Trim25 Is an RNA-Specific Activator of Lin28a/TuT4-Mediated Uridylation.
A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway.
Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms.
Janecki DM, Sen R, Szóstak N, Kajdasz A, Kordyś M, Plawgo K, Pandakov D, Philips A, Warkocki Z
Nucleic acids research 2024 Apr 12;52(6):3327-3345
Nucleic acids research 2024 Apr 12;52(6):3327-3345
TENT2, TUT4, and TUT7 selectively regulate miRNA sequence and abundance.
Yang A, Bofill-De Ros X, Stanton R, Shao TJ, Villanueva P, Gu S
Nature communications 2022 Sep 7;13(1):5260
Nature communications 2022 Sep 7;13(1):5260
Knockouts of TUT7 and 3'hExo show that they cooperate in histone mRNA maintenance and degradation.
Holmquist CE, He W, Meganck RM, Marzluff WF
RNA (New York, N.Y.) 2022 Nov;28(11):1519-1533
RNA (New York, N.Y.) 2022 Nov;28(11):1519-1533
RNA uridyl transferases TUT4/7 differentially regulate miRNA variants depending on the cancer cell type.
Medhi R, Price J, Furlan G, Gorges B, Sapetschnig A, Miska EA
RNA (New York, N.Y.) 2022 Mar;28(3):353-370
RNA (New York, N.Y.) 2022 Mar;28(3):353-370
Extracellular microRNA 3' end modification across diverse body fluids.
Koyano K, Bahn JH, Xiao X
Epigenetics 2021 Sep;16(9):1000-1015
Epigenetics 2021 Sep;16(9):1000-1015
Terminal uridyltransferase 7 regulates TLR4-triggered inflammation by controlling Regnase-1 mRNA uridylation and degradation.
Lin CC, Shen YR, Chang CC, Guo XY, Young YY, Lai TY, Yu IS, Lee CY, Chuang TH, Tsai HY, Hsu LC
Nature communications 2021 Jun 29;12(1):3878
Nature communications 2021 Jun 29;12(1):3878
YB1 regulates miR-205/200b-ZEB1 axis by inhibiting microRNA maturation in hepatocellular carcinoma.
Liu X, Chen D, Chen H, Wang W, Liu Y, Wang Y, Duan C, Ning Z, Guo X, Otkur W, Liu J, Qi H, Liu X, Lin A, Xia T, Liu HX, Piao HL
Cancer communications (London, England) 2021 Jul;41(7):576-595
Cancer communications (London, England) 2021 Jul;41(7):576-595
Identification of RNA-binding proteins that partner with Lin28a to regulate Dnmt3a expression.
Parisi S, Castaldo D, Piscitelli S, D'Ambrosio C, Divisato G, Passaro F, Avolio R, Castellucci A, Gianfico P, Masullo M, Scaloni A, Russo T
Scientific reports 2021 Jan 27;11(1):2345
Scientific reports 2021 Jan 27;11(1):2345
MEF2C shapes the microtranscriptome during differentiation of skeletal muscles.
Piasecka A, Sekrecki M, Szcześniak MW, Sobczak K
Scientific reports 2021 Feb 10;11(1):3476
Scientific reports 2021 Feb 10;11(1):3476
Critical Roles of Translation Initiation and RNA Uridylation in Endogenous Retroviral Expression and Neural Differentiation in Pluripotent Stem Cells.
Takahashi K, Jeong D, Wang S, Narita M, Jin X, Iwasaki M, Perli SD, Conklin BR, Yamanaka S
Cell reports 2020 Jun 2;31(9):107715
Cell reports 2020 Jun 2;31(9):107715
AGO-bound mature miRNAs are oligouridylated by TUTs and subsequently degraded by DIS3L2.
Yang A, Shao TJ, Bofill-De Ros X, Lian C, Villanueva P, Dai L, Gu S
Nature communications 2020 Jun 2;11(1):2765
Nature communications 2020 Jun 2;11(1):2765
A Mechanism for microRNA Arm Switching Regulated by Uridylation.
Kim H, Kim J, Yu S, Lee YY, Park J, Choi RJ, Yoon SJ, Kang SG, Kim VN
Molecular cell 2020 Jun 18;78(6):1224-1236.e5
Molecular cell 2020 Jun 18;78(6):1224-1236.e5
A programmed wave of uridylation-primed mRNA degradation is essential for meiotic progression and mammalian spermatogenesis.
Morgan M, Kabayama Y, Much C, Ivanova I, Di Giacomo M, Auchynnikava T, Monahan JM, Vitsios DM, Vasiliauskaitė L, Comazzetto S, Rappsilber J, Allshire RC, Porse BT, Enright AJ, O'Carroll D
Cell research 2019 Mar;29(3):221-232
Cell research 2019 Mar;29(3):221-232
Exonuclease requirements for mammalian ribosomal RNA biogenesis and surveillance.
Pirouz M, Munafò M, Ebrahimi AG, Choe J, Gregory RI
Nature structural & molecular biology 2019 Jun;26(6):490-500
Nature structural & molecular biology 2019 Jun;26(6):490-500
3' Uridylation Confers miRNAs with Non-canonical Target Repertoires.
Yang A, Bofill-De Ros X, Shao TJ, Jiang M, Li K, Villanueva P, Dai L, Gu S
Molecular cell 2019 Aug 8;75(3):511-522.e4
Molecular cell 2019 Aug 8;75(3):511-522.e4
Uridylation by TUT4/7 Restricts Retrotransposition of Human LINE-1s.
Warkocki Z, Krawczyk PS, Adamska D, Bijata K, Garcia-Perez JL, Dziembowski A
Cell 2018 Sep 6;174(6):1537-1548.e29
Cell 2018 Sep 6;174(6):1537-1548.e29
MiRAR-miRNA Activity Reporter for Living Cells.
Turk MA, Chung CZ, Manni E, Zukowski SA, Engineer A, Badakhshi Y, Bi Y, Heinemann IU
Genes 2018 Jun 19;9(6)
Genes 2018 Jun 19;9(6)
3' Uridylation controls mature microRNA turnover during CD4 T-cell activation.
Gutiérrez-Vázquez C, Enright AJ, Rodríguez-Galán A, Pérez-García A, Collier P, Jones MR, Benes V, Mizgerd JP, Mittelbrunn M, Ramiro AR, Sánchez-Madrid F
RNA (New York, N.Y.) 2017 Jun;23(6):882-891
RNA (New York, N.Y.) 2017 Jun;23(6):882-891
mRNA 3' uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome.
Morgan M, Much C, DiGiacomo M, Azzi C, Ivanova I, Vitsios DM, Pistolic J, Collier P, Moreira PN, Benes V, Enright AJ, O'Carroll D
Nature 2017 Aug 17;548(7667):347-351
Nature 2017 Aug 17;548(7667):347-351
Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs.
Łabno A, Warkocki Z, Kuliński T, Krawczyk PS, Bijata K, Tomecki R, Dziembowski A
Nucleic acids research 2016 Dec 1;44(21):10437-10453
Nucleic acids research 2016 Dec 1;44(21):10437-10453
Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4).
Thornton JE, Du P, Jing L, Sjekloca L, Lin S, Grossi E, Sliz P, Zon LI, Gregory RI
Nucleic acids research 2014 Oct;42(18):11777-91
Nucleic acids research 2014 Oct;42(18):11777-91
Trim25 Is an RNA-Specific Activator of Lin28a/TuT4-Mediated Uridylation.
Choudhury NR, Nowak JS, Zuo J, Rappsilber J, Spoel SH, Michlewski G
Cell reports 2014 Nov 20;9(4):1265-72
Cell reports 2014 Nov 20;9(4):1265-72
A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway.
Chang HM, Triboulet R, Thornton JE, Gregory RI
Nature 2013 May 9;497(7448):244-8
Nature 2013 May 9;497(7448):244-8
Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms.
Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI
Cell 2011 Nov 23;147(5):1066-79
Cell 2011 Nov 23;147(5):1066-79
No comments: Submit comment
Supportive validation
- Submitted by
- Proteintech Group (provider)
- Main image

- Experimental details
- MCF7 cells were subjected to SDS PAGE followed by western blot with 18980-1-AP(ZCCHC11 antibody) at dilution of 1:500
- Sample type
- cell line
Supportive validation
- Submitted by
- Proteintech Group (provider)
- Main image
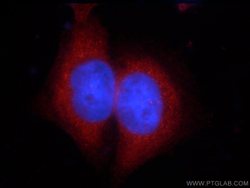
- Experimental details
- The ZCCHC11 antibody from Proteintech is a rabbit polyclonal antibody to a peptide of human ZCCHC11. This antibody recognizes human antigen. The ZCCHC11 antibody has been validated for the following applications: ELISA, WB, IF analysis.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot ELISA
ELISA