Antibody data
- Antibody Data
- Antigen structure
- References [3]
- Comments [0]
- Validations
- Immunohistochemistry [1]
- Other assay [2]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 14-6400-37 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- Smooth Muscle Myosin Monoclonal Antibody (SMMS-1), eBioscience™
- Antibody type
- Monoclonal
- Antigen
- Other
- Description
- Description: This SMMS-1 monoclonal antibody reacts with both isoforms of the human smooth muscle myosin heavy chain (SM1 and SM2) structural proteins, which are components of the contractile apparatus in smooth muscle cells. Smooth muscle myosin heavy chains are expressed by myoepithelial cells and also serve as the terminal differentiation marker for smooth muscle cells. In normal breast tissue, myoepithelial cells compose the outer layer of the lumen lining. In benign and in situ breast lesions, the intact myoepithelial layer stains with SMMS-1 and the loss of this staining indicates invasive cancer. This SMMS-1 antibody has also been reported to cross-react with bovine, canine, feline, and rat smooth muscle myosin heavy chains. Applications Reported: This SMMS-1 antibody has been reported for use in western blotting, and immunohistochemical staining of formalin-fixed paraffin embedded tissue sections. Applications Tested: This SMMS-1 antibody has been tested by western blot on rat heart and immunohistochemistry on formalin-fixed paraffin embedded tissue (with IHC Antigen Retrieval Solution - Low pH (Product # 00-4955-58). This can be used at less than or equal to 5 µg/mL. It is recommended that the antibody be titrated for optimal perfomance in the assay of interest. Purity: Greater than 90%, as determined by SDS-PAGE. Aggregation: Less than 10%, as determined by HPLC. Filtration: 0.2 µm post-manufacturing filtered.
- Reactivity
- Human, Rat, Bovine, Canine, Feline
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- SMMS-1
- Vial size
- 2 mg
- Concentration
- 0.5 mg/mL
- Storage
- 4°C
Submitted references Loss of the serine protease HTRA1 impairs smooth muscle cells maturation.
NFAT5 Isoform C Controls Biomechanical Stress Responses of Vascular Smooth Muscle Cells.
Immunostaining patterns of myoepithelial cells in breast lesions: a comparison of CD10 and smooth muscle myosin heavy chain.
Klose R, Prinz A, Tetzlaff F, Weis EM, Moll I, Rodriguez-Vita J, Oka C, Korff T, Fischer A
Scientific reports 2019 Dec 3;9(1):18224
Scientific reports 2019 Dec 3;9(1):18224
NFAT5 Isoform C Controls Biomechanical Stress Responses of Vascular Smooth Muscle Cells.
Zappe M, Feldner A, Arnold C, Sticht C, Hecker M, Korff T
Frontiers in physiology 2018;9:1190
Frontiers in physiology 2018;9:1190
Immunostaining patterns of myoepithelial cells in breast lesions: a comparison of CD10 and smooth muscle myosin heavy chain.
Kalof AN, Tam D, Beatty B, Cooper K
Journal of clinical pathology 2004 Jun;57(6):625-9
Journal of clinical pathology 2004 Jun;57(6):625-9
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
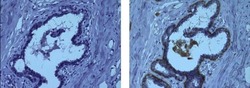
- Experimental details
- Immunohistochemistry on formalin-fixed paraffin embedded human infiltrating ductal carcinoma, using 5 µg/mL of Mouse IgG1 K Isotype Control Purified (Product # 14-4714-82) (left) or 5 µg/mL Anti-Smooth Muscle Myosin Purified (right) followed by Anti-Mouse IgG HRP, and DAB visualization.Nuclei are counterstained with hematoxylin.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
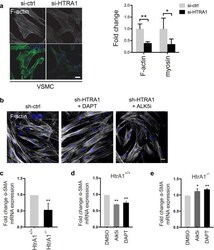
- Experimental details
- Figure 4 Contractile gene expression in HTRA1 -deficient VSMC is affected by Notch and TGFss signaling. ( a ) Representative photomicrographs of F-actin and immunolabeled myosin in HUASMC and quantification of F-actin and myosin positive areas (n = 4). ( b ) Representative photomicrographs of F-actin in HUASMC after four hours of TGFss inhibition using 10 uM ALK5 inhibitor (ALK5i, CAS 446859-33-2, Merck) or NOTCH3 inhibition for 16 hours using 25 uM DAPT (Merck) (n = 3). ( c - e ) Primary aortic VSMC derived from HtrA1 -/- or wildtype littermate controls were cultured until confluence was reached. Quantitative real-time PCR analysis of alpha smooth muscle actin (alpha-SMA). ( d , e ) VSMC were treated with DAPT (25 µM) for 16 hours or Alk5i (10 µM) for two hours before RNA isolation. Treatment with the solvent DMSO was used as control (n = 3). Statistical significance was determined by an unpaired student's t-test ( a,c,d,e ). *p < 0.05; **p < 0.01. Bar graphs show mean values, error bars indicate SD. Scale bar equals 10 um.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunohistochemistry
Immunohistochemistry