Antibody data
- Antibody Data
- Antigen structure
- References [3]
- Comments [0]
- Validations
- Flow cytometry [1]
- Other assay [3]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 14-1099-80 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- CD109 Monoclonal Antibody (HU17), eBioscience™
- Antibody type
- Monoclonal
- Antigen
- Other
- Description
- Description: The HU17 monoclonal antibody reacts with human CD109, also known as the platelet-specific Gov antigen. CD109, a GPI-anchored monomeric glycoprotein, exists in two isoforms of 120 and 150 kDa, which are proteolytically degraded from a 170 kDa form. CD109 is expressed on activated T cells, activated platelets and endothelial cells. CD109 expression is also found on a subset of CD34+ cells in adult and fetal bone marrow which contain megakaryocyte, myeloid, erythroid and multilineage progenitors, as well as pluriopotent hematopoietic stem cells. Recently, it has been demonstrated that CD109 is expressed on a variety of human cancer cell lines, and also lung and esophogeal cell carcinomas. In addition, CD109 was recently found to be a negative regulator of TGF-beta in keratinocytes. Applications Reported: This HU17 antibody has been reported for use in flow cytometric analysis, and immunoprecipitation. Applications Tested: This HU17 antibody has been tested by flow cytometric analysis of normal human peripheral blood. This can be used at less than or equal to 0.25 µg per test. A test is defined as the amount (µg) of antibody that will stain a cell sample in a final volume of 100 µL. Cell number should be determined empirically but can range from 10^5 to 10^8 cells/test. It is recommended that the antibody be carefully titrated for optimal performance in the assay of interest. Purity: Greater than 90%, as determined by SDS-PAGE. Aggregation: Less than 10%, as determined by HPLC. Filtration: 0.2 µm post-manufacturing filtered.
- Reactivity
- Human
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- HU17
- Vial size
- 25 μg
- Concentration
- 0.5 mg/mL
- Storage
- 4°C
Submitted references CD109 mediates tumorigenicity and cancer aggressiveness via regulation of EGFR and STAT3 signalling in cervical squamous cell carcinoma.
Mesotrypsin promotes malignant growth of breast cancer cells through shedding of CD109.
Identification of CD109 as part of the TGF-beta receptor system in human keratinocytes.
Mo XT, Leung TH, Tang HW, Siu MK, Wan PK, Chan KK, Cheung AN, Ngan HY
British journal of cancer 2020 Sep;123(5):833-843
British journal of cancer 2020 Sep;123(5):833-843
Mesotrypsin promotes malignant growth of breast cancer cells through shedding of CD109.
Hockla A, Radisky DC, Radisky ES
Breast cancer research and treatment 2010 Nov;124(1):27-38
Breast cancer research and treatment 2010 Nov;124(1):27-38
Identification of CD109 as part of the TGF-beta receptor system in human keratinocytes.
Finnson KW, Tam BY, Liu K, Marcoux A, Lepage P, Roy S, Bizet AA, Philip A
FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2006 Jul;20(9):1525-7
FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2006 Jul;20(9):1525-7
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
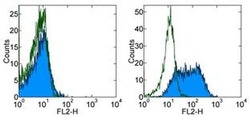
- Experimental details
- Staining of unstimulated (left) or 3-day PHA-stimulated (right) normal human peripheral blood cells with 0.125 µg of Mouse IgG1 kappa Isotype Control Purified (Product # 14-4714-82) (open histogram) or 0.125 µg of Anti-Human CD109 Purified (filled histogram) followed by F (ab')2 Anti-Mouse IgG PE (Product # 12-4012). Total viable cells were used for analysis.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
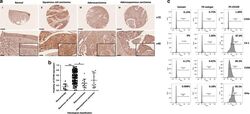
- Experimental details
- Fig. 1 Upregulation of CD109 was frequently detected in cervical squamous cell carcinoma. a Representative immunohistochemical CD109 staining of cervical cancer tissue microarray sections: weak CD109 staining in normal cervical tissue (i), strong CD109 staining in squamous cell carcinoma (ii), mild or moderate CD109 staining in adenocarcinoma (iii) and adenosquamous cell carcinoma (iv). Magnification: upper, x10; lower, x40. The insets highlight regions with x80 magnification. b Dot plot showing the distribution of CD109 immunoreactivity (presented as positivity, mean +- SD) in different histological classification groups. ** P 1 < 0.0001, * P 2 = 0.0007, Mann-Whitney test. c CD109( + ) subpopulation in cervical cancer cell lines C33A, C4-1, CaSki and SiHa was examined by flow cytometry.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
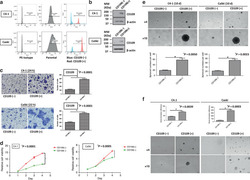
- Experimental details
- Fig. 2 Sorted CD109(+) cells displayed enhanced carcinogenic and aggressive abilities. The purity of CD109- expressed cells in sorted CD109(-) and CD109( + ) subpopulations was confirmed by flow-cytometric ( a ) and western blot analysis ( b ). c A representative image (left) and quantification bar graph (right) of migrated sorted CD109(-) and CD109( + ) subpopulations in C4-1 and CaSki cells (crystal violet). d Proliferation analyses of sorted CD109(-) and CD109( + ) subpopulations in C4-1 and CaSki cells. * P C4-1 < 0.0001, P CaSki = 0.0005, using the Student's t test. e Representative images of spheroids formed by sorted CD109(-) and CD109( + ) cells (above, magnification, x4, x10). Bar graph (below) represents the mean number of spheroids from each well under the microscope, and error bars represent SD, * P C4-1 = 0 . 005, P CaSki = 0.0023. f CD109- enhanced anchorage-independent cell growth ability was confirmed by soft-agar colony-formation assay. The sorted CD109( + ) cells formed larger and more clones than CD109(-) cells. * P C4-1 = 0 . 0039, P CaSki = 0.0003, using the Student's t test.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Fig. 4 Generation of CD109-CRISPR/Cas9 knockout clones. a The efficiency of the targeted deletion with CRISPR/CAS9 was determined by PCR in SiHa cells. Black arrows denote the cleaved band intensity of the transfected cells. b A schematic diagram depicting the CD109-guide RNA (gRNA2)-targeting region at exon 2 of CD109 (NC_000006.12) on chromosome 6; the PAM sequence is shown in red. The efficiency and precision of targeted knockout in CD109 were confirmed by TA-cloning sequencing analyses. TA-cloning DNA sequences of the wild-type (WT) and mutant sequences in KO clones were named Q4 and L65. Representative sequences for TA cloning are shown. Red boxes highlighted 1-base-pair (bp) insertion; the short black dash denotes 1-bp or 10-bp deletions. c Sequencing results of the predicted off-target genes (SPG11 and ARL8A) in Q4 and L65 cells. d The CD109 ( + ) subpopulation percentage in CD109 CRISPR/Cas9-knockout clones Q4 and L65 was examined by flow cytometry. e Western blot analysis for confirmation of CD109 CRISPR/Cas9 knockout in CaSki and SiHa cells.
 Explore
Explore Validate
Validate Learn
Learn Immunoprecipitation
Immunoprecipitation Flow cytometry
Flow cytometry