Antibody data
- Antibody Data
- Antigen structure
- References [5]
- Comments [0]
- Validations
- Flow cytometry [2]
- Other assay [3]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 16-9488-025 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- KLRG1 Monoclonal Antibody (13F12F2), Functional Grade, eBioscience™
- Antibody type
- Monoclonal
- Antigen
- Other
- Description
- Description: This 13F12F2 monoclonal antibody reacts with human killer cell lectin-like receptor subfamily G, member 1 (KLRG1), a type II transmembrane inhibitor receptor of the C-type lectin superfamily that contains an ITIM domain. This inhibitory receptor is expressed on subsets of gamma-delta T cells, NK (CD56dim), CD8+ and CD4+ T Cells. KLRG1 is expressed primarily by cells with an effector/memory phenotype that are short-lived, but capable of immediate effector cell function. Cadherin-E, -N, and -R are ligands for KLRG1. Cadherin/KLRG1 interaction inhibits cytolytic activity and proliferation. The percentage of KLRG1 positive cells can vary considerably, depending on antigen experience. The clones 13F12F2 and 13A2 appear to recognize a similar epitope based on cross-blocking studies. This 13F12F2 antibody has been tested by flow cytometric analysis of human peripheral blood cells. This can be used at less than or equal to 0.5 µg per test. A test is defined as the amount (µg) of antibody that will stain a cell sample in a final volume of 100 µL. Cell number should be determined empirically but can range from 10^5 to 10^8 cells/test. It is recommended that the antibody be carefully titrated for optimal performance in the assay of interest. Filtration: 0.2 µm post-manufacturing filtered. Purity: Greater than 90%, as determined by SDS-PAGE. Endotoxin Level: Less than 0.001 ng/µg antibody, as determined by LAL assay. Aggregation: Less than 10%, as determined by HPLC.
- Reactivity
- Human
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- 13F12F2
- Vial size
- 25 mg
- Concentration
- 1 mg/mL
- Storage
- 4°C, store in dark, DO NOT FREEZE!
Submitted references Immune cell phenotypes associated with disease severity and long-term neutralizing antibody titers after natural dengue virus infection.
Isolation of tumor-resident CD8(+) T cells from human lung tumors.
IL-1β, IL-23, and TGF-β drive plasticity of human ILC2s towards IL-17-producing ILCs in nasal inflammation.
Epithelial-mesenchymal transition leads to NK cell-mediated metastasis-specific immunosurveillance in lung cancer.
KLRG1 impairs CD4+ T cell responses via p16ink4a and p27kip1 pathways: role in hepatitis B vaccine failure in individuals with hepatitis C virus infection.
Rouers A, Chng MHY, Lee B, Rajapakse MP, Kaur K, Toh YX, Sathiakumar D, Loy T, Thein TL, Lim VWX, Singhal A, Yeo TW, Leo YS, Vora KA, Casimiro D, Lim B, Tucker-Kellogg L, Rivino L, Newell EW, Fink K
Cell reports. Medicine 2021 May 18;2(5):100278
Cell reports. Medicine 2021 May 18;2(5):100278
Isolation of tumor-resident CD8(+) T cells from human lung tumors.
Corgnac S, Lecluse Y, Mami-Chouaib F
STAR protocols 2021 Mar 19;2(1):100267
STAR protocols 2021 Mar 19;2(1):100267
IL-1β, IL-23, and TGF-β drive plasticity of human ILC2s towards IL-17-producing ILCs in nasal inflammation.
Golebski K, Ros XR, Nagasawa M, van Tol S, Heesters BA, Aglmous H, Kradolfer CMA, Shikhagaie MM, Seys S, Hellings PW, van Drunen CM, Fokkens WJ, Spits H, Bal SM
Nature communications 2019 May 14;10(1):2162
Nature communications 2019 May 14;10(1):2162
Epithelial-mesenchymal transition leads to NK cell-mediated metastasis-specific immunosurveillance in lung cancer.
Chockley PJ, Chen J, Chen G, Beer DG, Standiford TJ, Keshamouni VG
The Journal of clinical investigation 2018 Apr 2;128(4):1384-1396
The Journal of clinical investigation 2018 Apr 2;128(4):1384-1396
KLRG1 impairs CD4+ T cell responses via p16ink4a and p27kip1 pathways: role in hepatitis B vaccine failure in individuals with hepatitis C virus infection.
Shi L, Wang JM, Ren JP, Cheng YQ, Ying RS, Wu XY, Lin SM, Griffin JW, Li GY, Moorman JP, Yao ZQ
Journal of immunology (Baltimore, Md. : 1950) 2014 Jan 15;192(2):649-57
Journal of immunology (Baltimore, Md. : 1950) 2014 Jan 15;192(2):649-57
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
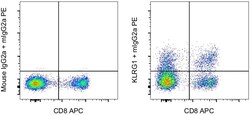
- Experimental details
- Normal human peripheral blood cells cells were stained with CD8 Monoclonal Antibody, APC (Product # 17-0088-42) and 0.25 µg of Mouse IgG2a kappa Isotype Control (Product # 14-4724-85) (left) or 0.25 µg of KLRG1 Monoclonal Antibody, Functional Grade (right) followed by IgG2a Monoclonal Antibody, PE (Product # 12-4210-82). Cells in the lymphocyte gate were used for analysis.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
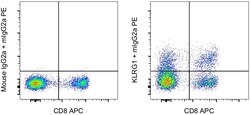
- Experimental details
- Normal human peripheral blood cells cells were stained with CD8 Monoclonal Antibody, APC (Product # 17-0088-42) and 0.25 µg of Mouse IgG2a kappa Isotype Control (Product # 14-4724-85) (left) or 0.25 µg of KLRG1 Monoclonal Antibody, Functional Grade (right) followed by IgG2a Monoclonal Antibody, PE (Product # 12-4210-82). Cells in the lymphocyte gate were used for analysis.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
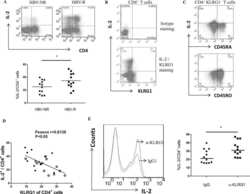
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Figure 2 Gating strategy for sorting CD8 + CD103 + and CD8 + KLRG1 + cells by FACS
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
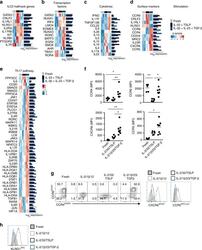
- Experimental details
- Fig. 6 ILC2 stimulations result in unique expression patterns. a Heatmap of genes related to ILC2 signature. b Heatmap of genes related to ILC-related transcription factors. c Heatmap of genes related to ILC-related cytokines. d Heatmap of genes encoding significantly differentially expressed surface markers d . Both z-scores (row-normalized) and absolute expression values are shown. e Heatmap of genes significantly different expressed in ILC2s upon stimulation with IL-2, IL-1beta, IL-23, and TGF-beta as compared with ILC2s cultured with IL-2, IL-33, and TSLP that are related to the Th17 KEGG pathway. f Quantification of expression of CCR4, CCR6, CCR5 and CXCR6 on freshly isolated blood ILC2s, and upon culture for 5 days with IL-2, IL-33 and TSLP or IL-2, IL-1beta, IL-23, and TGF-beta ( n = 5-8). g Representative flow cytometric analysis of CCR4, CCR6, CCR5, and CXCR6 upon culture as in f . h Representative flow cytometric analysis of KLRG1 expression upon culture as in f . Data are presented as individual values with mean of two to four independent experiments ( f ). * p < 0.05, ** p < 0.01 as determined by one way ANOVA. Source data
 Explore
Explore Validate
Validate Learn
Learn Flow cytometry
Flow cytometry