Antibody data
- Antibody Data
- Antigen structure
- References [2]
- Comments [0]
- Validations
- Immunocytochemistry [4]
- Immunohistochemistry [1]
- Other assay [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-49581 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- KMT2D Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Synthetic peptide
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 400 μL
- Concentration
- 0.5 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references The low-complexity domains of the KMT2D protein regulate histone monomethylation transcription to facilitate pancreatic cancer progression.
Cul4A-DDB1-mediated monoubiquitination of phosphoglycerate dehydrogenase promotes colorectal cancer metastasis via increased S-adenosylmethionine.
Li W, Wu L, Jia H, Lin Z, Zhong R, Li Y, Jiang C, Liu S, Zhou X, Zhang E
Cellular & molecular biology letters 2021 Nov 10;26(1):45
Cellular & molecular biology letters 2021 Nov 10;26(1):45
Cul4A-DDB1-mediated monoubiquitination of phosphoglycerate dehydrogenase promotes colorectal cancer metastasis via increased S-adenosylmethionine.
Zhang Y, Yu H, Zhang J, Gao H, Wang S, Li S, Wei P, Liang J, Yu G, Wang X, Li X, Li D, Yang W
The Journal of clinical investigation 2021 Nov 1;131(21)
The Journal of clinical investigation 2021 Nov 1;131(21)
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
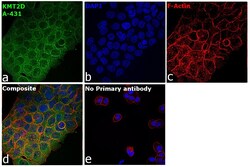
- Experimental details
- Immunofluorescence analysis of Histone-lysine N-methyltransferase 2D was performed using 70% confluent log phase A-431 cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 15 minutes, and blocked with 2% BSA for 45 minutes at room temperature. The cells were labeled with KMT2D Polyclonal Antibody (Product # PA5-49581) at 1:100 dilution in 0.1% BSA, incubated at 4 degree celsius overnight and then labeled with Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 (Product # A32790), (1:2000), for 45 minutes at room temperature (Panel a: Green). Nuclei (Panel b:Blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (Product # S36938). F-actin (Panel c: Red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image showing cytoplasmic localization. Panel e represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
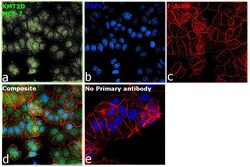
- Experimental details
- Immunofluorescence analysis of Histone-lysine N-methyltransferase 2D was performed using 70% confluent log phase MCF-7 cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 15 minutes, and blocked with 2% BSA for 45 minutes at room temperature. The cells were labeled with KMT2D Polyclonal Antibody (Product # PA5-49581) at 1:100 dilution in 0.1% BSA, incubated at 4 degree celsius overnight and then labeled with Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 (Product # A32790), (1:2000), for 45 minutes at room temperature (Panel a: Green). Nuclei (Panel b:Blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (Product # S36938). F-actin (Panel c: Red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image showing nuclear localization. Panel e represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
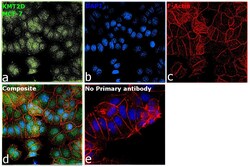
- Experimental details
- Immunofluorescence analysis of Histone-lysine N-methyltransferase 2D was performed using 70% confluent log phase MCF-7 cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 15 minutes, and blocked with 2% BSA for 45 minutes at room temperature. The cells were labeled with KMT2D Polyclonal Antibody (Product # PA5-49581) at 1:100 dilution in 0.1% BSA, incubated at 4 degree celsius overnight and then labeled with Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 (Product # A32790), (1:2000), for 45 minutes at room temperature (Panel a: Green). Nuclei (Panel b:Blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (Product # S36938). F-actin (Panel c: Red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image showing nuclear localization. Panel e represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
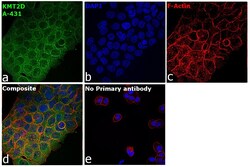
- Experimental details
- Immunofluorescence analysis of Histone-lysine N-methyltransferase 2D was performed using 70% confluent log phase A-431 cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 15 minutes, and blocked with 2% BSA for 45 minutes at room temperature. The cells were labeled with KMT2D Polyclonal Antibody (Product # PA5-49581) at 1:100 dilution in 0.1% BSA, incubated at 4 degree celsius overnight and then labeled with Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 (Product # A32790), (1:2000), for 45 minutes at room temperature (Panel a: Green). Nuclei (Panel b:Blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (Product # S36938). F-actin (Panel c: Red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image showing cytoplasmic localization. Panel e represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- Immunohistochemistry analysis of KMT2D in paraffin-embedded human brain tissue. Samples were incubated with KMT2D polyclonal antibody (Product # PA5-49581) using a dilution of 1:500 for 1 hour at room temperature followed by an undiluted biotinylated CRF Anti-Polyvalent HRP Polymer antibody.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
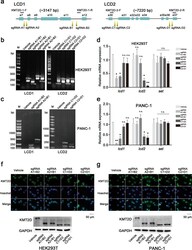
- Experimental details
- Fig. 2 LCD knockout strategy and verification in HEK293T and PANC-1 cells. a Schematic representation of LCD knockout strategy using CRSPR-Cas9 technology. LCD1 (located between exon 8 and 11) was deleted with sgRNA primers: two upstream primers (sgRNA-A1/A2) and two downstream primers (sgRNA-B1/B2). This was then verified by genotyping with KMT2D-1-F/R primers. For LCD2 (located between exon 31 and 36), two upstream primers (sgRNA-C1/C2) and two downstream primers (sgRNA-D1/D2) were used to delete LCD2 and then the rest was tested with KMT2D-2-F/R primers. b , c Verification with genotyping assays. In HEK293T and PANC-1 cells, genotyping PCR was performed in transfected cells. The results are consistent with the predictions compared with the control group (vehicle). d , e RT-PCR analyses to measure LCD knockout. lcd1 mRNA expression only significantly decreased in LCD1-deleted cells, while that of lcd2 only in LCD2-deleted cells. The set gene located at the C-terminal of the KMT2D gene was selected as a control (error bars denote standard deviation). f , g Representative images of immunofluorescences and western blotting in HEK293T and PANC-1 cells. After LCD knockout, the KMT2D protein level did not significantly decrease, and KMT2D LCD1 or KMT2D LCD2 was successfully deleted. KMT2D, 593 KDa; LCD1 deletion-KMT2D, 483 KDa; LCD2 deletion-KMT2D, 447 KDa. All the values are represented as means +- SD. Data are representative of three independent experiments
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry