Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Western blot [1]
- Immunohistochemistry [1]
- Other assay [2]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-31813 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- XPO6 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant full-length protein
- Description
- Recommended positive controls: IMR32, U87-MG. Predicted reactivity: Mouse (97%), Rat (97%). Store product as a concentrated solution. Centrifuge briefly prior to opening the vial.
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 1 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references Cancer-associated exportin-6 upregulation inhibits the transcriptionally repressive and anticancer effects of nuclear profilin-1.
Zhu C, Kim SJ, Mooradian A, Wang F, Li Z, Holohan S, Collins PL, Wang K, Guo Z, Hoog J, Ma CX, Oltz EM, Held JM, Shao J
Cell reports 2021 Feb 16;34(7):108749
Cell reports 2021 Feb 16;34(7):108749
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
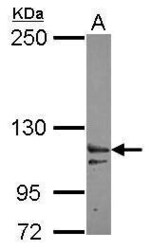
- Experimental details
- Western Blot using XPO6 Polyclonal Antibody (Product # PA5-31813). Sample (30 µg of whole cell lysate). Lane A: IMR32 . 5% SDS PAGE. XPO6 Polyclonal Antibody (Product # PA5-31813) diluted at 1:3,000.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
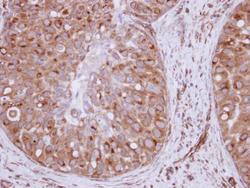
- Experimental details
- Immunohistochemical analysis of paraffin-embedded human breast cancer, using XPO6 (Product # PA5-31813) antibody at 1:250 dilution. Antigen Retrieval: EDTA based buffer, pH 8.0, 15 min.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Figure 1. XPO6 upregulation occurs in cancer and associates with poor patient survival (A) Pan-cancer XPO6 mRNA levels in the TCGA cohorts. Whiskers represent min-max. Mann-Whitney U non-parametric test was used to compare between normal and tumor samples for each cancer type. RSEM, RNA-seq by expectation maximization. (B) XPO6 mRNA levels of 112 breast tumors in the TCGA dataset with adjacent normal tissues. p value was based on Wilcoxon Signed Rank non-parametric test. (C) qRT-PCR of XPO6 mRNA levels in breast epithelial cell lines. One-way ANOVA and Dunnett''s multiple comparisons tests were used to compare between MCF-10A and breast cancer cell lines. Data are mean +- SEM. **p < 0.01; ***p < 0.001; ****p < 0.0001. (D) Western blot of XPO6 in untransformed and transformed breast epithelial cell lines, with different cytoplasmic and nuclear proteins as controls. (E) Univariate Kaplan-Meier analysis of the association between XPO6 mRNA levels and the overall survival (OS) of TCGA patients with bladder, renal clear cell, hepatocellular, and breast carcinomas. (F) Univariate Kaplan-Meier analysis of the association between XPO6 mRNA levels and the OS of stage II breast cancer patients within the TCGA cohort. (G) Univariate Kaplan-Meier analysis of the association between XPO6 protein levels and the OS of a cohort of 65 breast cancer patients. p values for (E)-(G) were based on log-rank tests. See also Figures S1 and S2 and Tables S1A - S1C .
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
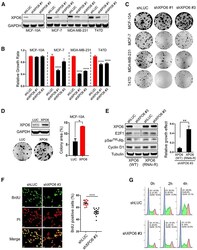
- Experimental details
- Figure 2. XPO6 is required for breast cancer cell growth in vitro (A) Human breast epithelial cell lines were infected with a small hairpin targeting luciferase (shLUC) and two different XPO6 shRNAs. (B) Relative growth effects of XPO6 knockdown by dividing normalized Alamar blue values (day 9/1) of shXPO6 versus shLUC cells. Data are mean +- SEM of a representative experiment (sextuples per condition). p values were based on one-way ANOVA and Dunnett''s multiple comparison tests. (C) Colony formation assay using cells in (A). (D) Colony formation assay using MCF-10A cells infected with XPO6 or luciferase. Colony areas were expressed as percentages. Data are mean +- SEM of a representative experiment (triplicates per condition). (E) MCF-7 cells expressing WT or RNAi-Res XPO6 were infected with shLUC or shXPO6 #3 and subjected to Western blot analysis. Lanes for the E2F1 blot were cropped and rearranged from the same blot (indicated by the black line). Relative growth effects of XPO6 knockdown were calculated by normalizing colony areas of shXPO6 versus shLUC cells. Data are mean +- SD of one representative experiment (triplicates per condition). (F) MCF-7 cells infected with shLUC or shXPO6 #3 were labeledwithBrdU and stained for BrdU or with propidium iodide (PI). Fifteen random fields were quantified. Percent BrdU positivity of cells in all images is shown. Data are mean +- SEM. Scale bars, 40 mum. (G) MCF-7 cells from (F) were synchronized by double thymidine block, releas
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot