Antibody data
- Antibody Data
- Antigen structure
- References [18]
- Comments [0]
- Validations
- Immunohistochemistry [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA035787 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA035787, RRID:AB_2674782
- Product name
- Anti-TMPRSS2
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human TMPRSS2, Gene description: transmembrane protease, serine 2, Alternative Gene Names: PRSS10, Validated applications: WB, IHC, Uniprot ID: O15393, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.2 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references ACE2 and TMPRSS2 in human kidney tissue and urine extracellular vesicles with age, sex, and COVID-19
A high-throughput screening system for SARS-CoV-2 entry inhibition, syncytia formation and cell toxicity
Resistance of Omicron subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to neutralizing antibodies
TMPRSS2 is a functional receptor for human coronavirus HKU1.
Angiotensin‐converting enzyme 2 and transmembrane protease serine 2 in female and male patients with end‐stage kidney disease
SARS-COV-2 spike binding to ACE2 in living cells monitored by TR-FRET
Impact of TMPRSS2 Expression, Mutation Prognostics, and Small Molecule (CD, AD, TQ, and TQFL12) Inhibition on Pan-Cancer Tumors and Susceptibility to SARS-CoV-2
Expanded ACE2 dependencies of diverse SARS-like coronavirus receptor binding domains
Systematic Investigation of SARS-CoV-2 Receptor Protein Distribution along Viral Entry Routes in Humans
The Spike-Stabilizing D614G Mutation Interacts with S1/S2 Cleavage Site Mutations To Promote the Infectious Potential of SARS-CoV-2 Variants.
Clinical and in Vitro Evidence against Placenta Infection at Term by Severe Acute Respiratory Syndrome Coronavirus 2
Tropism of Severe Acute Respiratory Syndrome Coronavirus 2 for Barrett’s Esophagus May Increase Susceptibility to Developing Coronavirus Disease 2019
Elevation in viral entry genes and innate immunity compromise underlying increased infectivity and severity of COVID-19 in cancer patients
Viral S protein histochemistry reveals few potential SARS-CoV-2 entry sites in human ocular tissues
SARS-CoV-2 Alpha, Beta, and Delta variants display enhanced Spike-mediated syncytia formation.
Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue.
Syncytia formation by SARS-CoV-2-infected cells.
SARS-CoV-2 Cell Entry Factors ACE2 and TMPRSS2 Are Expressed in the Microvasculature and Ducts of Human Pancreas but Are Not Enriched in β Cells.
Bach M, Laftih S, Andresen J, Pedersen R, Andersen T, Madsen L, Madsen K, Hinrichs G, Zachar R, Svenningsen P, Lund L, Johansen I, Hansen L, Palarasah Y, Jensen B
Pflügers Archiv - European Journal of Physiology 2024;477(1):83-98
Pflügers Archiv - European Journal of Physiology 2024;477(1):83-98
A high-throughput screening system for SARS-CoV-2 entry inhibition, syncytia formation and cell toxicity
Jancy S, Lupitha S, Chandrasekharan A, Varadarajan S, Nelson-Sathi S, Prasad R, Jones S, Easwaran S, Darvin P, Sivasailam A, Santhoshkumar T
Biological Procedures Online 2023;25(1)
Biological Procedures Online 2023;25(1)
Resistance of Omicron subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to neutralizing antibodies
Planas D, Bruel T, Staropoli I, Guivel-Benhassine F, Porrot F, Maes P, Grzelak L, Prot M, Mougari S, Planchais C, Puech J, Saliba M, Sahraoui R, Fémy F, Morel N, Dufloo J, Sanjuán R, Mouquet H, André E, Hocqueloux L, Simon-Loriere E, Veyer D, Prazuck T, Péré H, Schwartz O
Nature Communications 2023;14(1)
Nature Communications 2023;14(1)
TMPRSS2 is a functional receptor for human coronavirus HKU1.
Saunders N, Fernandez I, Planchais C, Michel V, Rajah MM, Baquero Salazar E, Postal J, Porrot F, Guivel-Benhassine F, Blanc C, Chauveau-Le Friec G, Martin A, Grzelak L, Oktavia RM, Meola A, Ahouzi O, Hoover-Watson H, Prot M, Delaune D, Cornelissen M, Deijs M, Meriaux V, Mouquet H, Simon-Lorière E, van der Hoek L, Lafaye P, Rey F, Buchrieser J, Schwartz O
Nature 2023 Dec;624(7990):207-214
Nature 2023 Dec;624(7990):207-214
Angiotensin‐converting enzyme 2 and transmembrane protease serine 2 in female and male patients with end‐stage kidney disease
Arefin S, Hernandez L, Ward L, Schwarz A, Barany P, Stenvinkel P, Kublickiene K
European Journal of Clinical Investigation 2022;52(8)
European Journal of Clinical Investigation 2022;52(8)
SARS-COV-2 spike binding to ACE2 in living cells monitored by TR-FRET
Cecon E, Burridge M, Cao L, Carter L, Ravichandran R, Dam J, Jockers R
Cell Chemical Biology 2022;29(1):74-83.e4
Cell Chemical Biology 2022;29(1):74-83.e4
Impact of TMPRSS2 Expression, Mutation Prognostics, and Small Molecule (CD, AD, TQ, and TQFL12) Inhibition on Pan-Cancer Tumors and Susceptibility to SARS-CoV-2
Fu J, Liu S, Tan Q, Liu Z, Qian J, Li T, Du J, Song B, Li D, Zhang L, He J, Guo K, Zhou B, Chen H, Fu S, Liu X, Cheng J, He T, Fu J
Molecules 2022;27(21):7413
Molecules 2022;27(21):7413
Expanded ACE2 dependencies of diverse SARS-like coronavirus receptor binding domains
Xie X, Roelle S, Shukla N, Pham A, Bruchez A, Matreyek K
PLOS Biology 2022;20(7):e3001738
PLOS Biology 2022;20(7):e3001738
Systematic Investigation of SARS-CoV-2 Receptor Protein Distribution along Viral Entry Routes in Humans
Bräutigam K, Reinhard S, Galván J, Wartenberg M, Hewer E, Schürch C
Respiration 2022;101(6):610-618
Respiration 2022;101(6):610-618
The Spike-Stabilizing D614G Mutation Interacts with S1/S2 Cleavage Site Mutations To Promote the Infectious Potential of SARS-CoV-2 Variants.
Gellenoncourt S, Saunders N, Robinot R, Auguste L, Rajah MM, Kervevan J, Jeger-Madiot R, Staropoli I, Planchais C, Mouquet H, Buchrieser J, Schwartz O, Chakrabarti LA
Journal of virology 2022 Oct 12;96(19):e0130122
Journal of virology 2022 Oct 12;96(19):e0130122
Clinical and in Vitro Evidence against Placenta Infection at Term by Severe Acute Respiratory Syndrome Coronavirus 2
Colson A, Depoix C, Dessilly G, Baldin P, Danhaive O, Hubinont C, Sonveaux P, Debiève F
The American Journal of Pathology 2021;191(9):1610-1623
The American Journal of Pathology 2021;191(9):1610-1623
Tropism of Severe Acute Respiratory Syndrome Coronavirus 2 for Barrett’s Esophagus May Increase Susceptibility to Developing Coronavirus Disease 2019
Jin R, Brown J, Li Q, Bayguinov P, Wang J, Mills J
Gastroenterology 2021;160(6):2165-2168.e4
Gastroenterology 2021;160(6):2165-2168.e4
Elevation in viral entry genes and innate immunity compromise underlying increased infectivity and severity of COVID-19 in cancer patients
Kwan J, Lin L, Bell R, Bruce J, Richardson C, Pugh T, Liu F
Scientific Reports 2021;11(1)
Scientific Reports 2021;11(1)
Viral S protein histochemistry reveals few potential SARS-CoV-2 entry sites in human ocular tissues
Martin G, Wolf J, Lapp T, Agostini H, Schlunck G, Auw-Hädrich C, Lange C
Scientific Reports 2021;11(1)
Scientific Reports 2021;11(1)
SARS-CoV-2 Alpha, Beta, and Delta variants display enhanced Spike-mediated syncytia formation.
Rajah MM, Hubert M, Bishop E, Saunders N, Robinot R, Grzelak L, Planas D, Dufloo J, Gellenoncourt S, Bongers A, Zivaljic M, Planchais C, Guivel-Benhassine F, Porrot F, Mouquet H, Chakrabarti LA, Buchrieser J, Schwartz O
The EMBO journal 2021 Dec 15;40(24):e108944
The EMBO journal 2021 Dec 15;40(24):e108944
Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue.
Aguiar JA, Tremblay BJ, Mansfield MJ, Woody O, Lobb B, Banerjee A, Chandiramohan A, Tiessen N, Cao Q, Dvorkin-Gheva A, Revill S, Miller MS, Carlsten C, Organ L, Joseph C, John A, Hanson P, Austin RC, McManus BM, Jenkins G, Mossman K, Ask K, Doxey AC, Hirota JA
The European respiratory journal 2020 Sep;56(3)
The European respiratory journal 2020 Sep;56(3)
Syncytia formation by SARS-CoV-2-infected cells.
Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, Planchais C, Porrot F, Guivel-Benhassine F, Van der Werf S, Casartelli N, Mouquet H, Bruel T, Schwartz O
The EMBO journal 2020 Dec 1;39(23):e106267
The EMBO journal 2020 Dec 1;39(23):e106267
SARS-CoV-2 Cell Entry Factors ACE2 and TMPRSS2 Are Expressed in the Microvasculature and Ducts of Human Pancreas but Are Not Enriched in β Cells.
Coate KC, Cha J, Shrestha S, Wang W, Gonçalves LM, Almaça J, Kapp ME, Fasolino M, Morgan A, Dai C, Saunders DC, Bottino R, Aramandla R, Jenkins R, Stein R, Kaestner KH, Vahedi G, HPAP Consortium, Brissova M, Powers AC
Cell metabolism 2020 Dec 1;32(6):1028-1040.e4
Cell metabolism 2020 Dec 1;32(6):1028-1040.e4
No comments: Submit comment
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Enhanced method
- Orthogonal validation
- Main image
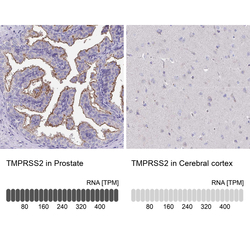
- Experimental details
- Immunohistochemistry analysis in human prostate and cerebral cortex tissues using HPA035787 antibody. Corresponding TMPRSS2 RNA-seq data are presented for the same tissues.
- Sample type
- Human
- Protocol
- Protocol
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunohistochemistry
Immunohistochemistry