Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Immunocytochemistry [4]
- Immunohistochemistry [1]
- Other assay [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 702593 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- DRD1 Recombinant Rabbit Monoclonal Antibody (1H8L2)
- Antibody type
- Monoclonal
- Antigen
- Synthetic peptide
- Description
- This antibody is predicted to react with Monkey, Cat, Bovine. Recombinant rabbit monoclonal antibodies are produced using in vitro expression systems. The expression systems are developed by cloning in the specific antibody DNA sequences from immunoreactive rabbits. Then, individual clones are screened to select the best candidates for production. The advantages of using recombinant rabbit monoclonal antibodies include: better specificity and sensitivity, lot-to-lot consistency, animal origin-free formulations, and broader immunoreactivity to diverse targets due to larger rabbit immune repertoire.
- Reactivity
- Human, Rat
- Host
- Rabbit
- Isotype
- IgG
- Antibody clone number
- 1H8L2
- Vial size
- 100 μg
- Concentration
- 0.5 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references Efficient and risk-reduced genome editing using double nicks enhanced by bacterial recombination factors in multiple species.
He X, Chen W, Liu Z, Yu G, Chen Y, Cai YJ, Sun L, Xu W, Zhong L, Gao C, Chen J, Zhang M, Yang S, Yao Y, Zhang Z, Ma F, Zhang CC, Lu HP, Yu B, Cheng TL, Qiu J, Sheng Q, Zhou HM, Lv ZR, Yan J, Zhou Y, Qiu Z, Cui Z, Zhang X, Meng A, Sun Q, Yang Y
Nucleic acids research 2020 Jun 4;48(10):e57
Nucleic acids research 2020 Jun 4;48(10):e57
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
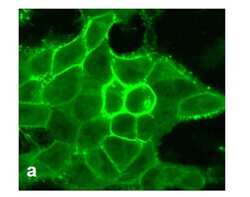
- Experimental details
- For immunofluorescence analysis, HEK-293 cells were stably transfected with human DRD1. After 24hr of transfection the cells were fixed and permeabilized for detection of endogenous DRD1 using Anti- DRD1 Recombinant Rabbit Monoclonal Antibody (Product # 702593, 1 µg/mL, 1:500 dilution) and labeled with Goat anti-Rabbit IgG (H+L) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate. Panel a) shows representative cells that were stained for detection of membrane localization of the DRD1 on the transfected cells. The images were captured at 40X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
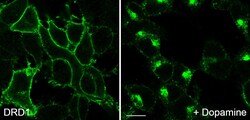
- Experimental details
- For immunocytochemistry analysis, HEK 293 cells stably transfected with Dopamine D1 receptor (D1R) were grown on poly-L-lysine-coated coverslips overnight. Cells were then treated with 10 µM dopamine for 30 min, washed with PBS and fixed with 4% paraformaldehyde and 0.2% picric acid in phosphate buffer (pH 6.9) for 30 min at room temperature and washed several times with phosphate buffer. Specimens were permeabilized with ice-cold methanol and then incubated with Anti- DRD1 Recombinant Rabbit Monoclonal Antibody (Product # 702593, 1 µg/mL). Cells were then incubated with Goat anti-Rabbit IgG (H+L) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate for 2 h at room temperature, mounted and examined using a laser scanning confocal microscope. Note, robust receptor internalization after agonist exposure.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
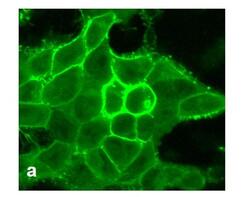
- Experimental details
- For immunofluorescence analysis, HEK-293 cells were stably transfected with human DRD1. After 24hr of transfection the cells were fixed and permeabilized for detection of endogenous DRD1 using Anti- DRD1 Recombinant Rabbit Monoclonal Antibody (Product # 702593, 1 µg/mL, 1:500 dilution) and labeled with Goat anti-Rabbit IgG (H+L) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate. Panel a) shows representative cells that were stained for detection of membrane localization of the DRD1 on the transfected cells. The images were captured at 40X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
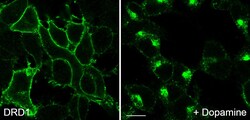
- Experimental details
- For immunocytochemistry analysis, HEK 293 cells stably transfected with Dopamine D1 receptor (D1R) were grown on poly-L-lysine-coated coverslips overnight. Cells were then treated with 10 µM dopamine for 30 min, washed with PBS and fixed with 4% paraformaldehyde and 0.2% picric acid in phosphate buffer (pH 6.9) for 30 min at room temperature and washed several times with phosphate buffer. Specimens were permeabilized with ice-cold methanol and then incubated with Anti- DRD1 Recombinant Rabbit Monoclonal Antibody (Product # 702593, 1 µg/mL). Cells were then incubated with Goat anti-Rabbit IgG (H+L) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate for 2 h at room temperature, mounted and examined using a laser scanning confocal microscope. Note, robust receptor internalization after agonist exposure.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
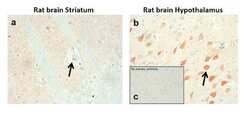
- Experimental details
- Sections of Rat brain striatum & hypothalamus were dewaxed, microwaved in citric acid and incubated with Anti-DRD1 Recombinant Rabbit Monoclonal Antibody (Product # 702593, 1 µg/mL, 1:500 dilution). Sections were then sequentially treated with biotinylated Anti-rabbit IgG and AB solution. Sections were further developed in AEC and lightly counterstained with hematoxylin. Panel a & b) shows the specific localization of DRD1 in neuronal stomata and dendrites in striatum and hypothalamus respectively. Panel c) shows the no primary antibody control.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
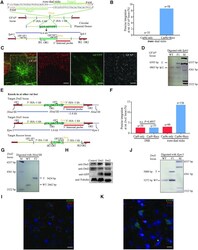
- Experimental details
- Figure 4. Trans -dual nicks with RecOFAR facilitates efficient gene KI in rats. ( A ) Schematic overview of the strategy to generate a GFAP-p2A-mEYFP KI allele. The nested PCR primers used for knock-in identification are shown ( Supplementary Table S4 ). Two targets are shown with green. The restriction endonuclease used for Southern blotting is SphI. A 3' internal probe on 3' homology arm is shown. ( B ) Germline transmission rate of p2A-mEYFP precise integration at the gfap locus without and with RecOFAR. ( C ) Immunostaining brain sections of gfap KI-positive F 1 rats. Neurons marked with NeuN are in red, mEYFP is in green and glial cells marked with anti-GFAP are in white. Scale bar in left panel is 200 mum. The region in the box is magnified in the right panels. Scale bar, 100 mum. ( D ) Southern blotting analysis of the gfap targeted allele and WT. T, KI target band, 6.6 kb. ( E ) Schematic overview of the strategy to generate the Drd2-p2A-ChR2-EYFP, Drd1-p2A-ChR2-EYFP and Bassoon -5.5 kb KI alleles. The restriction endonuclease HindIII and KpnI were used for Southern blotting at the Drd2 and Drd1 loci respectively. Two 3' internal probes on 3' homology arms are shown at the Drd2 and Drd1 loci. The PCR primers used for knock-in identification are shown ( Supplementary Table S4 ). ( F ) Germline transmission rate of p2A-ChR2-EYFP precise integration at the Drd2 locus with different strategies. ( G ) Southern blotting analysis of the Drd2-p2A-ChR2-EYFP targeted allele. (
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry