Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Immunocytochemistry [3]
- Immunohistochemistry [3]
- Other assay [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-64393 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- NCAPH2 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant protein fragment
- Description
- Immunogen sequence: EAENEFLSLD DFPDSRTNVD LKNDQTPSEV LIIPLLPMAL VAPDEMEKNN NPLYSRQGEV LASRKDFRMN TCVPHPRGAF MLEPEGMS Highest antigen sequence identity to the following orthologs: Mouse - 81%, Rat - 82%.
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 0.4 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references Cell division requires RNA eviction from condensing chromosomes.
Sharp JA, Perea-Resa C, Wang W, Blower MD
The Journal of cell biology 2020 Nov 2;219(11)
The Journal of cell biology 2020 Nov 2;219(11)
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
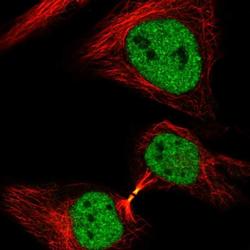
- Experimental details
- Immunofluorescent staining of NCAPH2 in human cell line HeLa shows positivity in cytokinetic bridge & nucleus but excluded from the nucleoli. Samples were probed using a NCAPH2 Polyclonal Antibody (Product # PA5-64393).
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
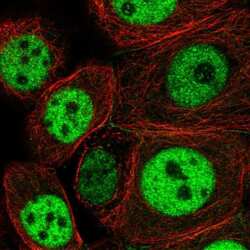
- Experimental details
- Immunofluorescent staining of NCAPH2 in human cell line MCF7 using a NCAPH2 Polyclonal Antibody (Product # PA5-64393) shows localization to nucleoplasm and cell junctions.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
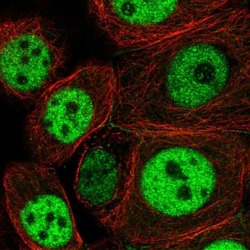
- Experimental details
- Immunofluorescent staining of NCAPH2 in human cell line MCF7 using a NCAPH2 Polyclonal Antibody (Product # PA5-64393) shows localization to nucleoplasm and cell junctions.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
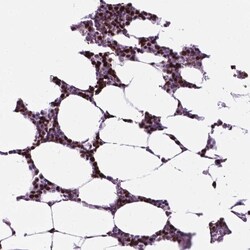
- Experimental details
- Immunohistochemical staining of NCAPH2 in human bone marrow using NCAPH2 Polyclonal Antibody (Product # PA5-64393) shows high expression.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
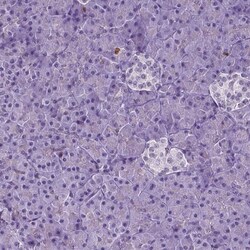
- Immunohistochemical staining of NCAPH2 in human pancreas using NCAPH2 Polyclonal Antibody (Product # PA5-64393) shows low expression as expected.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
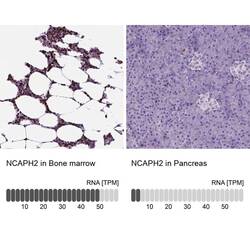
- Immunohistochemical staining of NCAPH2 in human bone marrow and pancreas tissues using NCAPH2 Polyclonal Antibody (Product # PA5-64393). Corresponding NCAPH2 RNA-seq data are presented for the same tissues.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
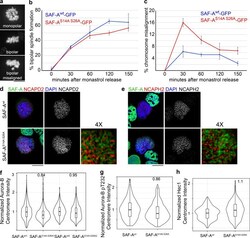
- Figure S5. Cells expressing phosphomutant SAF-A show a transient defect in kinetochore-microtubule error correction but show normal localization of mitotic factors. (a-c) To test for correction of kinetochore-microtubule attachment errors, SAF-A wt -GFP and SAF-A S14A S26A -GFP cells were treated with monastrol and subsequently released into media containing MG132. For each time point in the experiment, 100-300 cells were scored. (a) Examples of each chromosome alignment category scored after monastrol washout. (b) Quantitation of bipolar spindle assembly following monastrol washout. Error bars are the SD values from two experiments. (c) Quantitation of chromosome alignment following monastrol washout. We observed a 2.5-fold increase in chromosome misalignment in SAF-A S14A S26A -GFP cells 30 min after monastrol release. Error bars are the SD from two experiments. (d) Localization of condensin I subunit NCAPD2 in SAF-A wt -GFP and SAF-A S14A S26A -GFP cells. 4x magnification was produced using Adobe Photoshop. (e) Localization of condensin II subunit NCAPH2 in SAF-A wt -GFP and SAF-A S14A S26A -GFP cells. 4x magnification was produced in Photoshop. (f) Violin plot depicting normalized Aurora-B centromere intensity in SAF-A wt -GFP and SAF-A S14A S26A -GFP cells. For replicate 1, SAF-A wt1 ( n = 415 kinetochores), SAF-A S14A S26A1 ( n = 704 kinetochores). In replicate 2, SAF-A wt2 ( n = 1,447 kinetochores), SAF-A S14A S26A2 ( n = 1,927 kinetochores). (g) Violin plot depicting
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry