PA5-41830
antibody from Invitrogen Antibodies
Targeting: CPSF6
CFIM, CFIM68, CFIM72, HPBRII-4, HPBRII-7
Antibody data
- Antibody Data
- Antigen structure
- References [3]
- Comments [0]
- Validations
- Immunocytochemistry [3]
- Immunohistochemistry [1]
- Other assay [2]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-41830 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- CPSF6 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Synthetic peptide
- Description
- Peptide sequence: PPLAGPPNRG DRPPPPVLFP GQPFGQPPLG PLPPGPPPPV PGYGPPPGPP Sequence homology: Cow: 100%; Dog: 100%; Guinea Pig: 100%; Horse: 100%; Human: 100%; Mouse: 100%; Rabbit: 100%; Rat: 100%; Zebrafish: 79%
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 1 mg/mL
- Storage
- -20°C, Avoid Freeze/Thaw Cycles
Submitted references Localization and functions of native and eGFP-tagged capsid proteins in HIV-1 particles.
Suppression of premature transcription termination leads to reduced mRNA isoform diversity and neurodegeneration.
HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains.
Francis AC, Cereseto A, Singh PK, Shi J, Poeschla E, Engelman AN, Aiken C, Melikyan GB
PLoS pathogens 2022 Aug;18(8):e1010754
PLoS pathogens 2022 Aug;18(8):e1010754
Suppression of premature transcription termination leads to reduced mRNA isoform diversity and neurodegeneration.
LaForce GR, Farr JS, Liu J, Akesson C, Gumus E, Pinkard O, Miranda HC, Johnson K, Sweet TJ, Ji P, Lin A, Coller J, Philippidou P, Wagner EJ, Schaffer AE
Neuron 2022 Apr 20;110(8):1340-1357.e7
Neuron 2022 Apr 20;110(8):1340-1357.e7
HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains.
Francis AC, Marin M, Singh PK, Achuthan V, Prellberg MJ, Palermino-Rowland K, Lan S, Tedbury PR, Sarafianos SG, Engelman AN, Melikyan GB
Nature communications 2020 Jul 14;11(1):3505
Nature communications 2020 Jul 14;11(1):3505
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
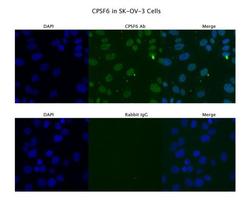
- Experimental details
- Immunofluorescence analysis of SKOV3 cells using an anti-CPSF6 polyclonal antibody (Product # PA5-41830). Primary Antibody Dilution: 4 µg/mL; Secondary Antibody: Anti-rabbit Alexa 546; Secondary Antibody Dilution: 2 µg/mL.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
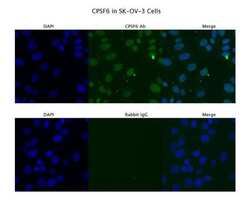
- Experimental details
- Immunofluorescence analysis of SKOV3 cells using an anti-CPSF6 polyclonal antibody (Product # PA5-41830). Primary Antibody Dilution: 4 µg/mL; Secondary Antibody: Anti-rabbit Alexa 546; Secondary Antibody Dilution: 2 µg/mL.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
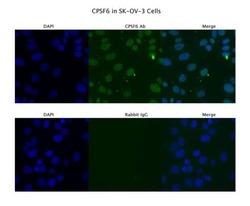
- Experimental details
- Immunofluorescence analysis of SKOV3 cells using an anti-CPSF6 polyclonal antibody (Product # PA5-41830). Primary Antibody Dilution: 4 µg/mL; Secondary Antibody: Anti-rabbit Alexa 546; Secondary Antibody Dilution: 2 µg/mL.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
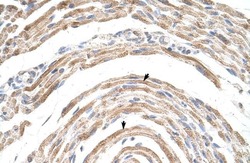
- Experimental details
- Immunohistochemistry (paraffin-embedded) analysis of human muscle tissue using an anti-CPSF6 polyclonal antibody (Product # PA5-41830).
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
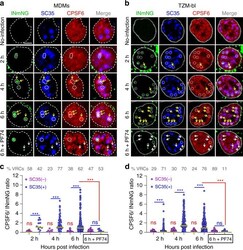
- Fig. 4 HIV-1 VRCs recruit CPSF6 to NSs. a - d Timecourse of CPSF6 accumulation at NS-localized HIV-1 complexes. TZM-bl cells stably expressing SNAP-Lamin nuclear envelope marker and MDMs (untreated) were infected at MOI of 5 and fixed at indicated time points. Where indicated, PF74 (25 muM) was added to samples 30 min before fixation (denoted 6 h + PF74). Cells were immunostained for NSs (SC35) and endogenous CPSF6. Uninfected cells were used as controls. Association of nuclear INmNG-labeled VRCs with SC35+ compartments in MDMs ( a ) and TZM-bl cells ( b ). White dashed circles and contours show NS-associated IN puncta, yellow arrows point to IN puncta that recruited CPSF6, white arrows in 6 h + PF74 treatment point to the loss of CPSF6 signal from IN puncta. Analysis of the ratio of CPSF6/INmNG fluorescence in nuclear IN puncta residing inside (SC35(+)) and outside (SC35(-)) NSs in MDMs ( c ) and TZM-bl cells ( d ). Note: raw INmNG and CPSF6 fluorescent intensities are shown in Supplementary Fig. 5a-d . Scale bar is 5 mum in ( a , b ). Data in ( c , d ) are presented as median values (yellow lines) +- SEM at 95% CI. Obtained by analysis of >80 nuclei in each cell type for each time point, from three independent experiments/donors. The number of MDM IN complexes analyzed in ( c ) was 31, 79, 353, and 80 for 2, 4, 6 h, and 6 h + PF74, respectively. In ( d ), >200 IN puncta in TZM-bl cells were analyzed for all time points. Statistical analysis was performed using nonparametric
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
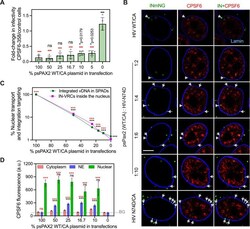
- 10.1371/journal.ppat.1010754.g006 Fig 6 HIV-1 nuclear import and integration targeting is associated with drastic changes to the structure and composition of the viral core. (A) The ability of HIV-1 with mixed WT/N74D CA to interact with cytoplasmic CPSF6-358 was determined in TZM-bl cells. The ratio of infection in CPSF6-358 to control permissive TZM-bl cells is plotted. Here, 5% WT/CA incorporated into N74D/CA mutant cores was sufficient for cytoplasmic CPSF6-358 interactions. The average values from 5 independent experiments with SEM are shown. (B) Single Z-plane images showing IN-labeled HIV-1 VRCs (green puncta) colocalized with CPSF6 in the nucleoplasm (yellow dashed lines) and VRCs stuck at the nuclear envelope without CPSF6 recruitment (white arrowheads) in fixed TZM-bl cells at 6 hpi. Endogenous CPSF6 (red) and the nuclear lamin (blue) were detected by immunostaining. Scale bar is 5 mum. (C) The efficiency of nuclear penetration of IN-labeled VRCs (magenta squares/ line) for viruses with mixed CA or a 100% N74D/CA was determined by normalizing to the nuclear penetration of 100% WT/CA at 6 hpi in TZM-bl cells. Nuclear penetration was defined by fraction of IN-VRCs detected inside >0.5 mum of the NE with respect to the total nucleus associated HIV-1 complexes (NE + inside nucleus). The efficiency of SPAD-targeted integration (green circles/line) of the same viral preps was determined in 293T cells after 5 days of infection by normalizing to the SPAD-localized integrati
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry