Antibody data
- Antibody Data
- Antigen structure
- References [10]
- Comments [0]
- Validations
- Western blot [1]
- Immunocytochemistry [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 21393-1-AP - Provider product page

- Provider
- Proteintech Group
- Proper citation
- Proteintech Cat#21393-1-AP, RRID:AB_10733883
- Product name
- RNF168 antibody
- Antibody type
- Polyclonal
- Description
- RNF168 antibody (Cat. #21393-1-AP) is a rabbit polyclonal antibody that shows reactivity with human and has been validated for the following applications: IP, WB, ELISA.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 20ul, 150ul
Submitted references A di-acetyl-decorated chromatin signature couples liquid condensation to suppress DNA end synapsis.
The HDAC6-RNF168 axis regulates H2A/H2A.X ubiquitination to enable double-strand break repair.
SENP1 Decreases RNF168 Phase Separation to Promote DNA Damage Repair and Drug Resistance in Colon Cancer.
A 1,2,3-Triazole Derivative of Quinazoline Exhibits Antitumor Activity by Tethering RNF168 to SQSTM1/P62.
Non-canonical function of DGCR8 in DNA double-strand break repair signaling and tumor radioresistance.
RNF8-ubiquitinated KMT5A is required for RNF168-induced H2A ubiquitination in response to DNA damage.
A feedforward circuit shaped by ECT2 and USP7 contributes to breast carcinogenesis.
An LTR retrotransposon-derived lncRNA interacts with RNF169 to promote homologous recombination.
GLP-catalyzed H4K16me1 promotes 53BP1 recruitment to permit DNA damage repair and cell survival.
p62-mediated Selective autophagy endows virus-transformed cells with insusceptibility to DNA damage under oxidative stress.
Bao K, Ma Y, Li Y, Shen X, Zhao J, Tian S, Zhang C, Liang C, Zhao Z, Yang Y, Zhang K, Yang N, Meng FL, Hao J, Yang J, Liu T, Yao Z, Ai D, Shi L
Molecular cell 2024 Apr 4;84(7):1206-1223.e15
Molecular cell 2024 Apr 4;84(7):1206-1223.e15
The HDAC6-RNF168 axis regulates H2A/H2A.X ubiquitination to enable double-strand break repair.
Qiu L, Xu W, Lu X, Chen F, Chen Y, Tian Y, Zhu Q, Liu X, Wang Y, Pei XH, Xu X, Zhang J, Zhu WG
Nucleic acids research 2023 Sep 22;51(17):9166-9182
Nucleic acids research 2023 Sep 22;51(17):9166-9182
SENP1 Decreases RNF168 Phase Separation to Promote DNA Damage Repair and Drug Resistance in Colon Cancer.
Wei M, Huang X, Liao L, Tian Y, Zheng X
Cancer research 2023 Sep 1;83(17):2908-2923
Cancer research 2023 Sep 1;83(17):2908-2923
A 1,2,3-Triazole Derivative of Quinazoline Exhibits Antitumor Activity by Tethering RNF168 to SQSTM1/P62.
Wang FC, Peng B, Ren TT, Liu SP, Du JR, Chen ZH, Zhang TT, Gu X, Li M, Cao SL, Xu X
Journal of medicinal chemistry 2022 Nov 24;65(22):15028-15047
Journal of medicinal chemistry 2022 Nov 24;65(22):15028-15047
Non-canonical function of DGCR8 in DNA double-strand break repair signaling and tumor radioresistance.
Hang Q, Zeng L, Wang L, Nie L, Yao F, Teng H, Deng Y, Yap S, Sun Y, Frank SJ, Chen J, Ma L
Nature communications 2021 Jun 29;12(1):4033
Nature communications 2021 Jun 29;12(1):4033
RNF8-ubiquitinated KMT5A is required for RNF168-induced H2A ubiquitination in response to DNA damage.
Lu X, Xu M, Zhu Q, Zhang J, Liu G, Bao Y, Gu L, Tian Y, Wen H, Zhu WG
FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2021 Apr;35(4):e21326
FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2021 Apr;35(4):e21326
A feedforward circuit shaped by ECT2 and USP7 contributes to breast carcinogenesis.
Zhang Q, Cao C, Gong W, Bao K, Wang Q, Wang Y, Bi L, Ma S, Zhao J, Liu L, Tian S, Zhang K, Yang J, Yao Z, Song N, Shi L
Theranostics 2020;10(23):10769-10790
Theranostics 2020;10(23):10769-10790
An LTR retrotransposon-derived lncRNA interacts with RNF169 to promote homologous recombination.
Deng B, Xu W, Wang Z, Liu C, Lin P, Li B, Huang Q, Yang J, Zhou H, Qu L
EMBO reports 2019 Nov 5;20(11):e47650
EMBO reports 2019 Nov 5;20(11):e47650
GLP-catalyzed H4K16me1 promotes 53BP1 recruitment to permit DNA damage repair and cell survival.
Lu X, Tang M, Zhu Q, Yang Q, Li Z, Bao Y, Liu G, Hou T, Lv Y, Zhao Y, Wang H, Yang Y, Cheng Z, Wen H, Liu B, Xu X, Gu L, Zhu WG
Nucleic acids research 2019 Dec 2;47(21):10977-10993
Nucleic acids research 2019 Dec 2;47(21):10977-10993
p62-mediated Selective autophagy endows virus-transformed cells with insusceptibility to DNA damage under oxidative stress.
Wang L, Howell MEA, Sparks-Wallace A, Hawkins C, Nicksic CA, Kohne C, Hall KH, Moorman JP, Yao ZQ, Ning S
PLoS pathogens 2019 Apr;15(4):e1007541
PLoS pathogens 2019 Apr;15(4):e1007541
No comments: Submit comment
Supportive validation
- Submitted by
- Proteintech Group (provider)
- Main image
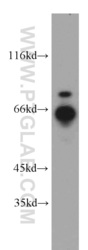
- Experimental details
- HepG2 cells were subjected to SDS PAGE followed by western blot with 21393-1-AP(RNF168 antibody) at dilution of 1:500
- Sample type
- cell line
Supportive validation
- Submitted by
- Proteintech Group (provider)
- Main image
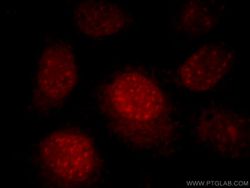
- Experimental details
- Immunofluorescent analysis of HepG2 cells, using RNF168 antibody 21393-1-AP at 1:25 dilution and Rhodamine-labeled goat anti-rabbit IgG (red).
- Sample type
- cell line
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot ELISA
ELISA