HPA027305
antibody from Atlas Antibodies
Targeting: NFKB1
KBF1, NF-kappaB, NF-kB1, NFkappaB, NFKB-p50, p105, p50
Antibody data
- Antibody Data
- Antigen structure
- References [4]
- Comments [0]
- Validations
- Western blot [1]
- Immunocytochemistry [1]
- Immunohistochemistry [1]
- Chromatin Immunoprecipitation [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA027305 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA027305, RRID:AB_1854429
- Product name
- Anti-NFKB1
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human NFKB1, Gene description: nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, Alternative Gene Names: KBF1, NF-kappaB, NF-kB1, NFkappaB, NFKB-p50, p105, p50, Validated applications: WB, IHC, ICC, ChIP, Uniprot ID: P19838, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.2 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references Multiple Nf1 Schwann cell populations reprogram the plexiform neurofibroma tumor microenvironment
MicroRNA Sequencing Analysis in Obstructive Sleep Apnea and Depression: Anti-Oxidant and MAOA-Inhibiting Effects of miR-15b-5p and miR-92b-3p through Targeting PTGS1-NF-κB-SP1 Signaling
Defining the structure of the NF-ĸB pathway in human immune cells using quantitative proteomic data.
Cancer-associated mesenchymal stroma fosters the stemness of osteosarcoma cells in response to intratumoral acidosis via NF-κB activation
Kershner L, Choi K, Wu J, Zhang X, Perrino M, Salomonis N, Shern J, Ratner N
JCI Insight 2022;7(18)
JCI Insight 2022;7(18)
MicroRNA Sequencing Analysis in Obstructive Sleep Apnea and Depression: Anti-Oxidant and MAOA-Inhibiting Effects of miR-15b-5p and miR-92b-3p through Targeting PTGS1-NF-κB-SP1 Signaling
Chen Y, Hsu P, Su M, Chen T, Hsiao C, Chin C, Liou C, Wang P, Wang T, Lin Y, Lee C, Lin M
Antioxidants 2021;10(11):1854
Antioxidants 2021;10(11):1854
Defining the structure of the NF-ĸB pathway in human immune cells using quantitative proteomic data.
Kok FO, Wang H, Riedlova P, Goodyear CS, Carmody RJ
Cellular signalling 2021 Dec;88:110154
Cellular signalling 2021 Dec;88:110154
Cancer-associated mesenchymal stroma fosters the stemness of osteosarcoma cells in response to intratumoral acidosis via NF-κB activation
Avnet S, Di Pompo G, Chano T, Errani C, Ibrahim-Hashim A, Gillies R, Donati D, Baldini N
International Journal of Cancer 2017;140(6):1331-1345
International Journal of Cancer 2017;140(6):1331-1345
No comments: Submit comment
Enhanced validation
- Submitted by
- Atlas Antibodies (provider)
- Enhanced method
- Genetic validation
- Main image

- Experimental details
- Western blot analysis in SK-BR-3 cells transfected with control siRNA, target specific siRNA probe #1 and #2, using Anti-NFKB1 antibody. Remaining relative intensity is presented. Loading control: Anti-GAPDH.
- Sample type
- Human
- Protocol
- Protocol
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
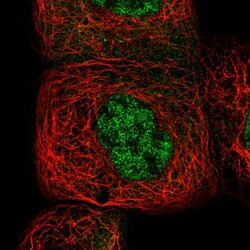
- Experimental details
- Immunofluorescent staining of human cell line A-431 shows localization to nucleoplasm & cytosol.
- Sample type
- Human
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Enhanced method
- Orthogonal validation
- Main image
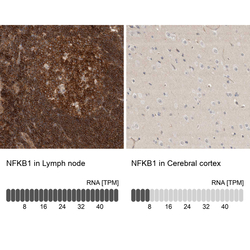
- Experimental details
- Immunohistochemistry analysis in human lymph node and cerebral cortex tissues using HPA027305 antibody. Corresponding NFKB1 RNA-seq data are presented for the same tissues.
- Sample type
- Human
- Protocol
- Protocol
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
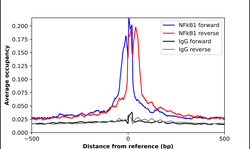
- Experimental details
- ChIP-Exo-Seq composite graph for Anti-NFKB1 (HPA027305, Lot 000014987) tested in K562 cells. Strand-specific reads (blue: forward, red: reverse) and IgG controls (black: forward, grey: reverse) are plotted against the distance from a composite set of reference binding sites. The antibody exhibits robust target enrichment compared to a non-specific IgG control and precisely reveals its structural organization around the binding site. Data generated by Prof. B. F. Pugh´s Lab at Cornell University.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry