Antibody data
- Antibody Data
- Antigen structure
- References [4]
- Comments [0]
- Validations
- Western blot [1]
- Immunocytochemistry [4]
- Immunohistochemistry [5]
- Chromatin Immunoprecipitation [1]
- Other assay [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA1-850 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- CREB Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Synthetic peptide
- Description
- PA1-850 detects both the phosphorylated and non-phosphorylated forms of cyclic-AMP response element binding protein (CREB) from rat, mouse, and human samples. PA1-850 has been successfully used in ChIP, Western blot, ICC/IF and IHC-P procedures. By Western blot, this antibody detects an ~43 kDa protein representing CREB from GH4 cell extract. The PA1-850 immunogen is a synthetic peptide corresponding to residues K(123) R R E I L S R R P S Y R K(136) of human CREB. This sequence is completely conserved between human, bovine, canine, mouse, pig, sheep, and rat CREB protein. The PA1-850 immunizing peptide (Cat. # PEP-142) is available for use in neutralization and control experiments.
- Reactivity
- Human, Mouse, Rat
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μg
- Concentration
- 1 mg/mL
- Storage
- -20°C, Avoid Freeze/Thaw Cycles
Submitted references microRNA-454-mediated NEDD4-2/TrkA/cAMP axis in heart failure: Mechanisms and cardioprotective implications.
Isx9 Regulates Calbindin D28K Expression in Pancreatic β Cells and Promotes β Cell Survival and Function.
Distinct cAMP response element-binding protein (CREB) domains stimulate different steps in a concerted mechanism of transcription activation.
Defective thymocyte proliferation and IL-2 production in transgenic mice expressing a dominant-negative form of CREB.
Wang Y, Pan W, Bai X, Wang X, Wang Y, Yin Y
Journal of cellular and molecular medicine 2021 Jun;25(11):5082-5098
Journal of cellular and molecular medicine 2021 Jun;25(11):5082-5098
Isx9 Regulates Calbindin D28K Expression in Pancreatic β Cells and Promotes β Cell Survival and Function.
Pujol JB, Heikkila E, Savoia C, Hajibeigi A, De Marchi U, Battiprolu PK, Öz OK, Dioum EHM
International journal of molecular sciences 2018 Aug 27;19(9)
International journal of molecular sciences 2018 Aug 27;19(9)
Distinct cAMP response element-binding protein (CREB) domains stimulate different steps in a concerted mechanism of transcription activation.
Kim J, Lu J, Quinn PG
Proceedings of the National Academy of Sciences of the United States of America 2000 Oct 10;97(21):11292-6
Proceedings of the National Academy of Sciences of the United States of America 2000 Oct 10;97(21):11292-6
Defective thymocyte proliferation and IL-2 production in transgenic mice expressing a dominant-negative form of CREB.
Barton K, Muthusamy N, Chanyangam M, Fischer C, Clendenin C, Leiden JM
Nature 1996 Jan 4;379(6560):81-5
Nature 1996 Jan 4;379(6560):81-5
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
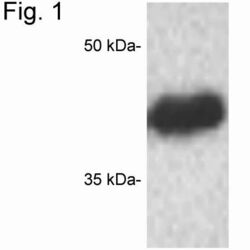
- Experimental details
- Western blot of CREB on GH4 cell extract using Product # PA1-850.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
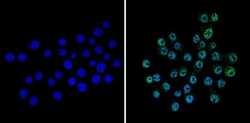
- Experimental details
- Immunofluorescent analysis of CREB (green) showing staining in the nucleus of Neuro-2a cells (right) compared to a negative control without primary antibody (left). Formalin-fixed cells were permeabilized with 0.1% Triton X-100 in TBS for 5-10 minutes and blocked with 3% BSA-PBS for 30 minutes at room temperature. Cells were probed with a CREB polyclonal antibody (Product # PA1-850) in 3% BSA-PBS at a dilution of 1:100 and incubated overnight at 4ºC in a humidified chamber. Cells were washed with PBST and incubated with a DyLight-conjugated secondary antibody in PBS at room temperature in the dark. Nuclei were stained with Hoechst or DAPI (blue). Images were taken at a magnification of 60x.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
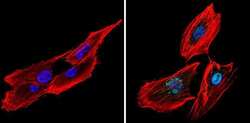
- Experimental details
- Immunofluorescent analysis of CREB (green) showing staining in the nucleus of SK-N-MC cells (right) compared to a negative control without primary antibody (left). Formalin-fixed cells were permeabilized with 0.1% Triton X-100 in TBS for 5-10 minutes and blocked with 3% BSA-PBS for 30 minutes at room temperature. Cells were probed with a CREB polyclonal antibody (Product # PA1-850) in 3% BSA-PBS at a dilution of 1:200 and incubated overnight at 4ºC in a humidified chamber. Cells were washed with PBST and incubated with a DyLight-conjugated secondary antibody in PBS at room temperature in the dark. Actin was stained using Alexa Fluor 554 (red) and nuclei were stained with Hoechst or DAPI (blue). Images were taken at a magnification of 60x.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
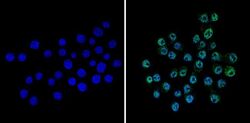
- Experimental details
- Immunofluorescent analysis of CREB (green) showing staining in the nucleus of Neuro-2a cells (right) compared to a negative control without primary antibody (left). Formalin-fixed cells were permeabilized with 0.1% Triton X-100 in TBS for 5-10 minutes and blocked with 3% BSA-PBS for 30 minutes at room temperature. Cells were probed with a CREB polyclonal antibody (Product # PA1-850) in 3% BSA-PBS at a dilution of 1:100 and incubated overnight at 4ºC in a humidified chamber. Cells were washed with PBST and incubated with a DyLight-conjugated secondary antibody in PBS at room temperature in the dark. Nuclei were stained with Hoechst or DAPI (blue). Images were taken at a magnification of 60x.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
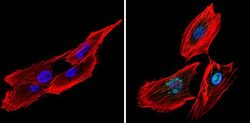
- Experimental details
- Immunofluorescent analysis of CREB (green) showing staining in the nucleus of SK-N-MC cells (right) compared to a negative control without primary antibody (left). Formalin-fixed cells were permeabilized with 0.1% Triton X-100 in TBS for 5-10 minutes and blocked with 3% BSA-PBS for 30 minutes at room temperature. Cells were probed with a CREB polyclonal antibody (Product # PA1-850) in 3% BSA-PBS at a dilution of 1:200 and incubated overnight at 4ºC in a humidified chamber. Cells were washed with PBST and incubated with a DyLight-conjugated secondary antibody in PBS at room temperature in the dark. Actin was stained using Alexa Fluor 554 (red) and nuclei were stained with Hoechst or DAPI (blue). Images were taken at a magnification of 60x.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
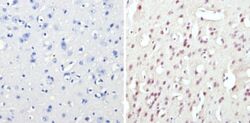
- Experimental details
- Immunohistochemistry analysis of CREB showing staining in the nucleus of paraffin-embedded mouse brain tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a CREB polyclonal antibody (Product # PA1-850) diluted in 3% BSA-PBS at a dilution of 1:50 overnight at 4°C in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
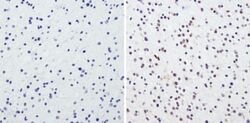
- Experimental details
- Immunohistochemistry analysis of CREB showing staining in the nucleus of paraffin-embedded human glioma (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a CREB polyclonal antibody (Product # PA1-850) diluted in 3% BSA-PBS at a dilution of 1:100 overnight at 4°C in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
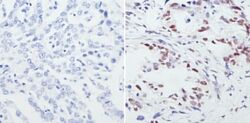
- Experimental details
- Immunohistochemistry analysis of CREB showing staining in the nucleus of paraffin-embedded human lung adenocarcinoma (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a CREB polyclonal antibody (Product # PA1-850) diluted in 3% BSA-PBS at a dilution of 1:100 overnight at 4°C in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
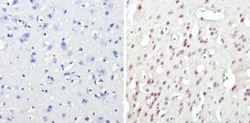
- Experimental details
- Immunohistochemistry analysis of CREB showing staining in the nucleus of paraffin-embedded mouse brain tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a CREB polyclonal antibody (Product # PA1-850) diluted in 3% BSA-PBS at a dilution of 1:50 overnight at 4°C in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
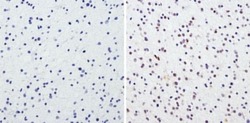
- Experimental details
- Immunohistochemistry analysis of CREB showing staining in the nucleus of paraffin-embedded human glioma (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a CREB polyclonal antibody (Product # PA1-850) diluted in 3% BSA-PBS at a dilution of 1:100 overnight at 4°C in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
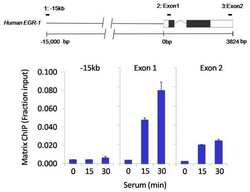
- Experimental details
- Chromatin immunoprecipitation analysis of CREB was performed using cross-linked chromatin from 1x10^6 HCT116 colon carcinoma cells treated with serum for 0, 15, and 30 minutes. Immunoprecipitation was performed using a multiplex microplate Matrix ChIP assay (see reference for Matrix ChIP protocol: http://www.ncbi.nlm.nih.gov/pubmed/22098709) with 1.0 µL/100 µL well volume of a CREB polyclonal antibody (Product # PA1-850). Chromatin aliquots from ~1x10^5 cells were used per ChIP pull-down. Quantitative PCR data were done in quadruplicate using 1 µL of eluted DNA in 2 µL SYBR real-time PCR reactions containing primers to amplify -15kb upstream of the Egr1 gene or exon-1 or exon-2 of Egr1. PCR calibration curves were generated for each primer pair from a dilution series of sheared total genomic DNA. Quantitation of immunoprecipitated chromatin is presented as signal relative to the total amount of input chromatin. Results represent the mean +/- SEM for three experiments. A schematic representation of the Egr-1 locus is shown above the data where boxes represent exons (black boxes = translated regions, white boxes = untranslated regions), the zigzag line represents an intron, and the straight line represents upstream sequence. Regions amplified by Egr-1 primers are represented by black bars. Data courtesy of the Innovators Program.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
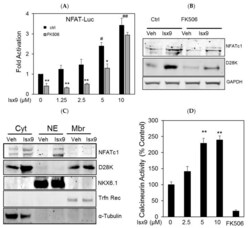
- Experimental details
- Figure 2 Isx9 increased NFAT transcriptional activity. ( A ) Dose-dependent activation of the NFAT reporter in beta cells after 24 h treatment with increasing dose of Isx9 in the presence or absence of 0.3 uM FK506, mean +- SEM of three independent experiments in triplicates, # p < 0.05 and ## p < 0.01 Isx9 versus non-treated cells; * p < 0.05, ** p < 0.01 effect of FK506 treatment versus control for each Isx9 dose. ( B ) Immunoblotting of NFATc1 and D28K in MIN6 whole cell extract after 48 h treatment with vehicle DMSO (Veh) or 10 uM Isx9 in the presence or absence of calcineurin inhibitor FK506. ( C ) Subcellular fractionation (Pierce) of MIN6 cells treated with Isx9 or vehicle into cytoplasmic (Cyt), nuclear (NE) and membrane (Mbr) fractions followed by immunoblotting of NFATc1 and D28K. alpha-Tubulin, Nkx6.1, and Transferrin receptors are used as loading controls. ( D ) Calcineurin activity in MIN6 represented as % of untreated cells treated with increasing doses of Isx9, FK506 is used as a negative control, mean +- SEM of three independent experiments in triplicates, ** p < 0.01 vs. control. ( E ) Immunoblotting of phospho-Creb1-Ser 133, D28K, and GAPDH after increasing dose of Isx9 for 24 h or ( F ) after 8 h and 24 h treatment with 10 uM Isx9 in MIN6 cells.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot