14-9326-37
antibody from Invitrogen Antibodies
Targeting: EPCAM
17-1A, 323/A3, CD326, CO-17A, EGP-2, EGP34, EGP40, Ep-CAM, ESA, GA733-2, HEA125, KS1/4, KSA, Ly74, M4S1, MH99, MIC18, MK-1, MOC31, TACST-1, TACSTD1, TROP1
 Western blot
Western blot Immunocytochemistry
Immunocytochemistry Immunoprecipitation
Immunoprecipitation Immunohistochemistry
Immunohistochemistry Flow cytometry
Flow cytometry Other assay
Other assayAntibody data
- Antibody Data
- Antigen structure
- References [22]
- Comments [0]
- Validations
- Immunocytochemistry [4]
- Immunohistochemistry [2]
- Flow cytometry [2]
- Other assay [21]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 14-9326-37 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- CD326 (EpCAM) Monoclonal Antibody (1B7), eBioscience™
- Antibody type
- Monoclonal
- Antigen
- Other
- Description
- Description: EpCAM (Epithelial cell adhesion molecule, CD326, KSA, TROP1) is a 40 kD cell-surface adhesion molecule participating in homophilic, calcium-independent cell-cell interactions. EpCAM is a type-I transmembrane protein, and is expressed primarily on the basolateral surface of most epithelia. Although normal epithelia express low levels of EpCAM, increased expression has been correlated with increased proliferation and progression to a mesenchymal phenotype. EpCAM has also been used as a diagnostic marker on circulating metastatic carcinoma cells, while cancer cells of non-epithelial origin do not express EpCAM. Applications Reported: This 1B7 antibody has been reported for use in flow cytometric analysis, immunohistochemistry of formalin-fixed paraffin embedded or frozen tissue sections, and immunoprecipitation. The 1B7 antibody has also been found useful for immunoblotting, and recognizes a protein of approximately 40 kD under non-reducing conditions. Applications Tested: This 1B7 antibody has been tested by flow cytometric analysis of A549 human lung carcinoma cells. This can be used at less than or equal to 0.25 µg per test. A test is defined as the amount (µg) of antibody that will stain a cell sample in a final volume of 100 µL. Cell number should be determined empirically but can range from 10^5 to 10^8 cells/test. The 1B7 antibody has been tested by immunohistochemistry of formalin-fixed paraffin embedded human tissue using low or high pH antigen retrieval and can be used at less than or equal to 10 µg/mL. It is recommended that the antibody be carefully titrated for optimal performance in the assay of interest. Purity: Greater than 90%, as determined by SDS-PAGE. Aggregation: Less than 10%, as determined by HPLC. Filtration: 0.2 µm post-manufacturing filtered.
- Reactivity
- Human
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- 1B7
- Vial size
- 2 mg
- Concentration
- 0.5 mg/mL
- Storage
- 4°C
Submitted references Selection of Cancer Stem Cell-Targeting Agents Using Bacteriophage Display.
The sodium/glucose cotransporters as potential therapeutic targets for CF lung diseases revealed by human lung organoid swelling assay.
Parathyroid Hormone-Related Protein Inhibition Blocks Triple-Negative Breast Cancer Expansion in Bone Through Epithelial to Mesenchymal Transition Reversal.
EMP3 negatively modulates breast cancer cell DNA replication, DNA damage repair, and stem-like properties.
Human Nasal and Lung Tissues Infected Ex Vivo with SARS-CoV-2 Provide Insights into Differential Tissue-Specific and Virus-Specific Innate Immune Responses in the Upper and Lower Respiratory Tract.
Chemotherapeutic Stress Influences Epithelial-Mesenchymal Transition and Stemness in Cancer Stem Cells of Triple-Negative Breast Cancer.
Helicobacter pylori promote inflammation and host defense through the cagA-dependent activation of mTORC1.
FZD5 contributes to TNBC proliferation, DNA damage repair and stemness.
Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity.
Triple-negative breast cancer targeting and killing by EpCAM-directed, plasmonically active nanodrug systems.
A genome editing approach to study cancer stem cells in human tumors.
Evidence of In Vitro Preservation of Human Nephrogenesis at the Single-Cell Level.
TGF-β1 secreted by Tregs in lymph nodes promotes breast cancer malignancy via up-regulation of IL-17RB.
Stage-specific embryonic antigen-3 (SSEA-3) and β3GalT5 are cancer specific and significant markers for breast cancer stem cells.
A lung-on-a-chip array with an integrated bio-inspired respiration mechanism.
The isolation and characterization of CTC subsets related to breast cancer dormancy.
Clostridium perfringens enterotoxin C-terminal domain labeled to fluorescent dyes for in vivo visualization of micrometastatic chemotherapy-resistant ovarian cancer.
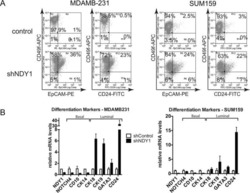
NDY1/KDM2B functions as a master regulator of polycomb complexes and controls self-renewal of breast cancer stem cells.
GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression.
Estrogen and progesterone together expand murine endometrial epithelial progenitor cells.
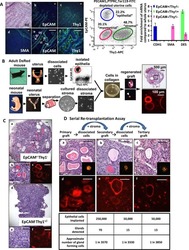
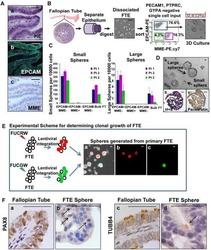
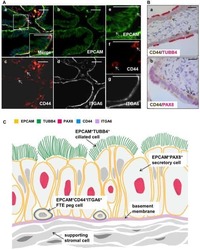
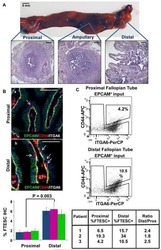
Stem-like epithelial cells are concentrated in the distal end of the fallopian tube: a site for injury and serous cancer initiation.
Slow-cycling therapy-resistant cancer cells.
Prater AR, Deutscher SL
Methods in molecular biology (Clifton, N.J.) 2022;2394:787-810
Methods in molecular biology (Clifton, N.J.) 2022;2394:787-810
The sodium/glucose cotransporters as potential therapeutic targets for CF lung diseases revealed by human lung organoid swelling assay.
Hirai H, Liang X, Sun Y, Zhang Y, Zhang J, Chen YE, Mou H, Zhao Y, Xu J
Molecular therapy. Methods & clinical development 2022 Mar 10;24:11-19
Molecular therapy. Methods & clinical development 2022 Mar 10;24:11-19
Parathyroid Hormone-Related Protein Inhibition Blocks Triple-Negative Breast Cancer Expansion in Bone Through Epithelial to Mesenchymal Transition Reversal.
Li J, Camirand A, Zakikhani M, Sellin K, Guo Y, Luan X, Mihalcioiu C, Kremer R
JBMR plus 2022 Jun;6(6):e10587
JBMR plus 2022 Jun;6(6):e10587
EMP3 negatively modulates breast cancer cell DNA replication, DNA damage repair, and stem-like properties.
Zhou K, Sun Y, Dong D, Zhao C, Wang W
Cell death & disease 2021 Sep 12;12(9):844
Cell death & disease 2021 Sep 12;12(9):844
Human Nasal and Lung Tissues Infected Ex Vivo with SARS-CoV-2 Provide Insights into Differential Tissue-Specific and Virus-Specific Innate Immune Responses in the Upper and Lower Respiratory Tract.
Alfi O, Yakirevitch A, Wald O, Wandel O, Izhar U, Oiknine-Djian E, Nevo Y, Elgavish S, Dagan E, Madgar O, Feinmesser G, Pikarsky E, Bronstein M, Vorontsov O, Jonas W, Ives J, Walter J, Zakay-Rones Z, Oberbaum M, Panet A, Wolf DG
Journal of virology 2021 Jun 24;95(14):e0013021
Journal of virology 2021 Jun 24;95(14):e0013021
Chemotherapeutic Stress Influences Epithelial-Mesenchymal Transition and Stemness in Cancer Stem Cells of Triple-Negative Breast Cancer.
Li X, Strietz J, Bleilevens A, Stickeler E, Maurer J
International journal of molecular sciences 2020 Jan 8;21(2)
International journal of molecular sciences 2020 Jan 8;21(2)
Helicobacter pylori promote inflammation and host defense through the cagA-dependent activation of mTORC1.
Feng GJ, Chen Y, Li K
Journal of cellular physiology 2020 Dec;235(12):10094-10108
Journal of cellular physiology 2020 Dec;235(12):10094-10108
FZD5 contributes to TNBC proliferation, DNA damage repair and stemness.
Sun Y, Wang Z, Na L, Dong D, Wang W, Zhao C
Cell death & disease 2020 Dec 12;11(12):1060
Cell death & disease 2020 Dec 12;11(12):1060
Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity.
Nguyen QH, Pervolarakis N, Blake K, Ma D, Davis RT, James N, Phung AT, Willey E, Kumar R, Jabart E, Driver I, Rock J, Goga A, Khan SA, Lawson DA, Werb Z, Kessenbrock K
Nature communications 2018 May 23;9(1):2028
Nature communications 2018 May 23;9(1):2028
Triple-negative breast cancer targeting and killing by EpCAM-directed, plasmonically active nanodrug systems.
Jenkins SV, Nima ZA, Vang KB, Kannarpady G, Nedosekin DA, Zharov VP, Griffin RJ, Biris AS, Dings RPM
NPJ precision oncology 2017;1(1):27
NPJ precision oncology 2017;1(1):27
A genome editing approach to study cancer stem cells in human tumors.
Cortina C, Turon G, Stork D, Hernando-Momblona X, Sevillano M, Aguilera M, Tosi S, Merlos-Suárez A, Stephan-Otto Attolini C, Sancho E, Batlle E
EMBO molecular medicine 2017 Jul;9(7):869-879
EMBO molecular medicine 2017 Jul;9(7):869-879
Evidence of In Vitro Preservation of Human Nephrogenesis at the Single-Cell Level.
Pode-Shakked N, Gershon R, Tam G, Omer D, Gnatek Y, Kanter I, Oriel S, Katz G, Harari-Steinberg O, Kalisky T, Dekel B
Stem cell reports 2017 Jul 11;9(1):279-291
Stem cell reports 2017 Jul 11;9(1):279-291
TGF-β1 secreted by Tregs in lymph nodes promotes breast cancer malignancy via up-regulation of IL-17RB.
Huang SC, Wei PC, Hwang-Verslues WW, Kuo WH, Jeng YM, Hu CM, Shew JY, Huang CS, Chang KJ, Lee EY, Lee WH
EMBO molecular medicine 2017 Dec;9(12):1660-1680
EMBO molecular medicine 2017 Dec;9(12):1660-1680
Stage-specific embryonic antigen-3 (SSEA-3) and β3GalT5 are cancer specific and significant markers for breast cancer stem cells.
Cheung SK, Chuang PK, Huang HW, Hwang-Verslues WW, Cho CH, Yang WB, Shen CN, Hsiao M, Hsu TL, Chang CF, Wong CH
Proceedings of the National Academy of Sciences of the United States of America 2016 Jan 26;113(4):960-5
Proceedings of the National Academy of Sciences of the United States of America 2016 Jan 26;113(4):960-5
A lung-on-a-chip array with an integrated bio-inspired respiration mechanism.
Stucki AO, Stucki JD, Hall SR, Felder M, Mermoud Y, Schmid RA, Geiser T, Guenat OT
Lab on a chip 2015 Mar 7;15(5):1302-10
Lab on a chip 2015 Mar 7;15(5):1302-10
The isolation and characterization of CTC subsets related to breast cancer dormancy.
Vishnoi M, Peddibhotla S, Yin W, T Scamardo A, George GC, Hong DS, Marchetti D
Scientific reports 2015 Dec 3;5:17533
Scientific reports 2015 Dec 3;5:17533
Clostridium perfringens enterotoxin C-terminal domain labeled to fluorescent dyes for in vivo visualization of micrometastatic chemotherapy-resistant ovarian cancer.
Cocco E, Shapiro EM, Gasparrini S, Lopez S, Schwab CL, Bellone S, Bortolomai I, Sumi NJ, Bonazzoli E, Nicoletti R, Deng Y, Saltzman WM, Zeiss CJ, Centritto F, Black JD, Silasi DA, Ratner E, Azodi M, Rutherford TJ, Schwartz PE, Pecorelli S, Santin AD
International journal of cancer 2015 Dec 1;137(11):2618-29
International journal of cancer 2015 Dec 1;137(11):2618-29
NDY1/KDM2B functions as a master regulator of polycomb complexes and controls self-renewal of breast cancer stem cells.
Kottakis F, Foltopoulou P, Sanidas I, Keller P, Wronski A, Dake BT, Ezell SA, Shen Z, Naber SP, Hinds PW, McNiel E, Kuperwasser C, Tsichlis PN
Cancer research 2014 Jul 15;74(14):3935-46
Cancer research 2014 Jul 15;74(14):3935-46
GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression.
Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z
Nature cell biology 2013 Feb;15(2):201-13
Nature cell biology 2013 Feb;15(2):201-13
Estrogen and progesterone together expand murine endometrial epithelial progenitor cells.
Janzen DM, Cheng D, Schafenacker AM, Paik DY, Goldstein AS, Witte ON, Jaroszewicz A, Pellegrini M, Memarzadeh S
Stem cells (Dayton, Ohio) 2013 Apr;31(4):808-22
Stem cells (Dayton, Ohio) 2013 Apr;31(4):808-22
Stem-like epithelial cells are concentrated in the distal end of the fallopian tube: a site for injury and serous cancer initiation.
Paik DY, Janzen DM, Schafenacker AM, Velasco VS, Shung MS, Cheng D, Huang J, Witte ON, Memarzadeh S
Stem cells (Dayton, Ohio) 2012 Nov;30(11):2487-97
Stem cells (Dayton, Ohio) 2012 Nov;30(11):2487-97
Slow-cycling therapy-resistant cancer cells.
Moore N, Houghton J, Lyle S
Stem cells and development 2012 Jul 1;21(10):1822-30
Stem cells and development 2012 Jul 1;21(10):1822-30
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
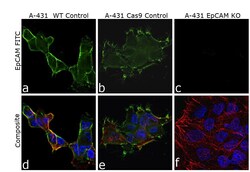
- Experimental details
- Knockout of EpCAM was achieved by CRISPR-Cas9 genome editing using LentiArray™ Lentiviral sgRNA (Product # A32042, AssayID CRISPR701274_LV) and LentiArray Cas9 Lentivirus (Product # A32064). Immunofluorescence analysis was performed on wild type A-431 cells (panel a,d), A-431 Cas9 control cells (panel b,e) and A-431 EpCAMKO cells (panel c, f). Cells were fixed, permeabilized, and labeled with EpCAM Monoclonal Antibody (Ber-EP4) (Product # 14-9326-82, 5 µg/mL dilution), followed by Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 488 (Product # A32723, 1:2000 dilution). Nuclei (blue) were stained using ProLong™ Diamond Antifade Mountant with DAPI (Product # P36962), and Rhodamine Phalloidin (Product # R415, 1:300) was used for cytoskeletal F-actin (red) staining. Loss of signal (panel c,f) upon CRISPR mediated knockout (KO) confirms that antibody is specific to EpCAM (green). The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
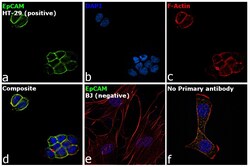
- Experimental details
- Immunofluorescence analysis of EpCAM was performed using HT-29 and BJ cells. The cells were fixed with 4% paraformaldehyde for 10 minutes and blocked with 2% BSA for 1 hour at room temperature. The cells were labeled with EpCAM Mouse Monoclonal Antibody (Product # 14-9326-82) at 5 µg/mL in 0.1% BSA and incubated overnight at 4 degree and then labeled with Goat anti-Mouse IgG (H+L) Superclonal™ Recombinant Secondary Antibody, Alexa Fluor® 488 (Product # A28175) at a dilution of 1:2000 for 45 minutes at room temperature (Panel a: green) in HT-29 cells. Nuclei (Panel b: blue) were stained with ProLong™ Diamond Antifade Mountant with DAPI (Product # P36962). F-actin (Panel c: red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image of HT-29 cells, which is a positive model for EpCAM expression showing a plasma membrane localization. Panel e represents the merged image of BJ cells, that are null for EpCAM protein expression. Panel f represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Immunofluorescence analysis of EpCAM was performed using HT-29 and BJ cells. The cells were fixed with 4% paraformaldehyde for 10 minutes and blocked with 2% BSA for 1 hour at room temperature. The cells were labeled with EpCAM Mouse Monoclonal Antibody (Product # 14-9326-82) at 5 µg/mL in 0.1% BSA and incubated overnight at 4 degree and then labeled with Goat anti-Mouse IgG (H+L) Superclonal™ Recombinant Secondary Antibody, Alexa Fluor® 488 (Product # A28175) at a dilution of 1:2000 for 45 minutes at room temperature (Panel a: green) in HT-29 cells. Nuclei (Panel b: blue) were stained with ProLong™ Diamond Antifade Mountant with DAPI (Product # P36962). F-actin (Panel c: red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image of HT-29 cells, which is a positive model for EpCAM expression showing a plasma membrane localization. Panel e represents the merged image of BJ cells, that are null for EpCAM protein expression. Panel f represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Knockout of EpCAM was achieved by CRISPR-Cas9 genome editing using LentiArray™ Lentiviral sgRNA (Product # A32042, AssayID CRISPR701274_LV) and LentiArray Cas9 Lentivirus (Product # A32064). Immunofluorescence analysis was performed on wild type A-431 cells (panel a,d), A-431 Cas9 control cells (panel b,e) and A-431 EpCAMKO cells (panel c, f). Cells were fixed, permeabilized, and labeled with EpCAM Monoclonal Antibody (Ber-EP4) (Product # 14-9326-82, 5 µg/mL dilution), followed by Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 488 (Product # A32723, 1:2000 dilution). Nuclei (blue) were stained using ProLong™ Diamond Antifade Mountant with DAPI (Product # P36962), and Rhodamine Phalloidin (Product # R415, 1:300) was used for cytoskeletal F-actin (red) staining. Loss of signal (panel c,f) upon CRISPR mediated knockout (KO) confirms that antibody is specific to EpCAM (green). The images were captured at 60X magnification.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
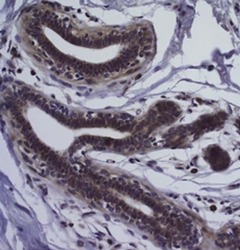
- Experimental details
- Immunohistochemistry of formalin-fixed paraffin embedded human breast tissue using 10 µg/mL Anti-Human CD326 (EpCAM) Purified followed by Anti-Mouse IgG Biotin, Avidin HRP, and DAB visualization (right).Nuclei are counterstained with hematoxyin.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
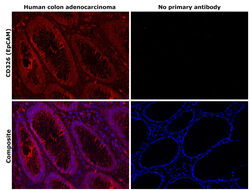
- Experimental details
- Immunohistochemical analysis of CD326 (EpCAM) was performed using formalin-fixed paraffin-embedded human colon adenocarcinoma tissue sections. To expose the target protein, heat-induced epitope retrieval was performed on de-paraffinized sections using eBioscience™ IHC Antigen Retrieval Solution - Low pH (10X) (Product # 00-4955-58) diluted to 1X solution in water in a decloaking chamber at 110 degree Celsius for 15 minutes. Following antigen retrieval, the sections were blocked with 3% H2O2 for 1 h at room temperature followed by 2% normal goat serum in 1X PBS for 45 minutes at room temperature. The sections were then probed with or without CD326 (EpCAM) Monoclonal Antibody (1B7), eBioscience™ (Product # 14-9326-82) at a concentration of 1 µg/mL in 0.1% normal goat serum overnight at 4 degree Celsius in a humidified chamber. Detection was performed using Alexa Fluor™ 647 Tyramide SuperBoost™ Kit, goat anti-mouse IgG (Product # B40916). Nuclei were stained with DAPI (Product # D1306) and the sections were mounted using ProLong™ Glass Antifade Mountant (Product # P36984). The images were captured on EVOS™ M7000 Imaging System (Product # AMF7000) at 20X magnification and externally deconvoluted.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
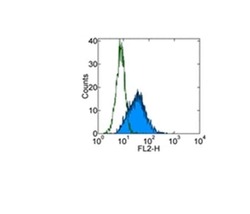
- Experimental details
- Staining of A549 human lung carcinoma cells with 0.125 µg of Purified Mouse IgG1 K Isotype Control (Product # 14-4714-82) (open histogram) or 0.125 µg of Anti-Human CD326 (EpCAM) Purified (filled histogram) followed by F (ab')2 Anti-Mouse IgG PE (Product # 12-4012). Total viable cells were used for analysis.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Staining of A549 human lung carcinoma cells with 0.125 µg of Purified Mouse IgG1 K Isotype Control (Product # 14-4714-82) (open histogram) or 0.125 µg of Anti-Human CD326 (EpCAM) Purified (filled histogram) followed by F (ab')2 Anti-Mouse IgG PE (Product # 12-4012). Total viable cells were used for analysis.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
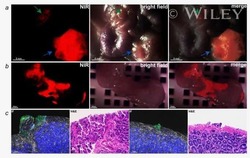
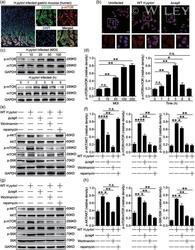
- 2 Figure H. pylori activate PI3K/AKT/mTOR/S6K pathway in gastric epithelial cells in a cagA -dependent manner. (a) Representative immunofluorescence staining images showing p-mTOR-expressing (red) CD326 + gastric epithelial cells (green) in gastric mucosa of H. pylori -infected patients. Scale bar = 100 um. (b) Representative immunofluorescence staining images showing p-mTOR expression in WT H. pylori -infected, DeltacagA -infected, and uninfected AGS cells. Red, p-mTOR; blue, DAPI-stained nuclei. Scale bar = 10 um. (c and d) Representative data (c) and statistical analysis (d) of p-mTOR and mTOR proteins in H. pylori -infected and uninfected AGS cells at a different time point (MOI = 100) or with different MOI (3 hr) analyzed by western blot were shown ( n = 3). (e and f) Representative data (e) and statistical analysis (f) of p-AKT and AKT proteins, p-mTOR and mTOR proteins, p-S6K and S6K proteins in WT H. pylori -infected, DeltacagA -infected, and uninfected AGS cells (MOI = 100, 3 hr) or in WT H. pylori -infected AGS cells (MOI = 100, 3 hr) pretreated with Wortmannin or rapamycin analyzed by western blot were shown ( n = 3). (e and f) Representative data (e) and statistical analysis (f) of p-AKT and AKT proteins, p-mTOR and mTOR proteins, p-S6K and S6K proteins in WT H. pylori -infected, DeltacagA -infected, and uninfected human primary gastric epithelial cells (MOI = 100, 3 hr) or in WT H. pylori -infected human primary gastric epithelial cells (MOI = 100, 3 hr) pretreat
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
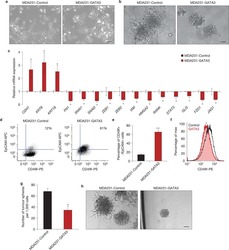
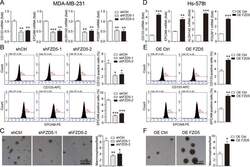
- Fig. 5 FZD5 maintains stem cell-like properties. A CD133, EPCAM, ALDH1A2, and POU5F1 expression in MDA-MB-231 cells was detected by real-time PCR. B Fractions of CD133-positive and EPCAM-positive MDA-MB-231 cells were detected by flow cytometry. C Mammosphere formation of MDA-MB-231 cells was shown. Scale bar: 500 mum, mean +- SD, n = 3. * P < 0.05, ** P < 0.01, *** P < 0.001, vs. shCtrl. D CD133, EPCAM, ALDH1A2, and POU5F1 expression in Hs-578t cells was detected by real-time PCR. E Fractions of CD133-positive and E PCAM-positive Hs-578t cells were detected by flow cytometry. F Mammosphere formation of Hs-578t cells was shown. Scale bar: 500 mum, mean +- SD, n = 3. * P < 0.05, ** P < 0.01, *** P < 0.001, vs. OE Ctrl.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
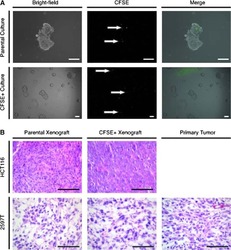
- Figure 1 Multiparametric flow cytometry of PBMCs capturing uPAR/int beta1 CTC subsets. Breast cancer PBMCs were first sorted applying gating parameters to select for DAPI - (4', 6-diamidino-2-phenylindole)/EpCAM - /CD45 - /CD44 + /CD24 - cells. Cells were then subsequently sorted to obtain uPAR/int beta1 subsets containing combinatorial expression of these markers. Antibodies used for flow cytometry and cell sorting were: anti-human CD45-APC-Cy7 (Biolegend, cat # 304015, 1:50 dilution), mouse anti-human EpCAM-PE CD326 (eBiosciences, cat # 12-9326-71, 1:40 dilution), anti-human CD24-PE ML5 (Biolegend, cat # 311106, 1:20 dilution), anti-human CD44-PE-Cy7 IM7 (Biolegend, cat # 103030, 1:20 dilution), mouse anti-human uPAR (CD87)-FITC (AbD Serotec cat # MCA2506FT, 1:10 dilution), anti-human int beta1 (CD29)-ApC TS2/16 (Biolegend, cat # 3030008, 1:50 dilution). Cells were confirmed to be CTCs by performing RT-PCR, immunoflurescence and genotyping arrays. Representative images are shown.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
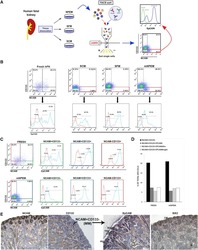
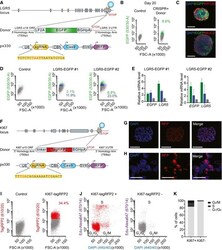
- Figure 1 LGR 5- EGFP and KI 67-Tag RFP 2 knock-in PDO s Design of LGR5-EGFP donor and CRISPR/Cas9 sgRNA vectors. Blue circle represents the CRISPR/Cas9 protein complex and the yellow box underneath the guide RNA. Flow cytometry profiles at day 20 post-nucleofection. Immunofluorescence for DAPI, EGFP, and KRT20 or MUC2 in in vitro cultured PDO#7-LGR5-EGFP#1. Scale bars indicate 100 mum. FACS profiles showing EGFP-high (green), -low (blue), and -negative (gray) cells in PDO#7-LGR5-EGFP#1 and #2 organoids. Relative mRNA expression level by real-time qPCR in cells expressing distinct levels of EGFP isolated from PDO#7-LGR5-EGFP#1 and #2 knock-in organoids. Values show mean +- s.d. of three measurements. Design of KI67-TagRFP2 donor and CRISPR/Cas9 sgRNA vectors. Blue circle represents the CRISPR/Cas9 protein complex and the yellow box underneath the guide RNA. Images of PDO#7-KI67-TagRFP2#1 organoids. Scale bars indicate 100 mum. PDO#7-KI67-TagRFP2#1 xenograft. TagRFP2 co-localizes with DAPI nuclear staining. Scale bars indicate 25 mum. Flow cytometry analysis of EPCAM + /DAPI - cell population of PDO#7-LGR5-EGFP/KI67-TagRFP2#1 from disaggregated xenografts. Cell cycle analysis of KI67-TagRFP2-positive and KI67-TagRFP2-negative cells from PDO#7-LGR5-EGFP/KI67-TagRFP2#1 disaggregated xenografts. X -axis shows DNA content and y -axis EdU incorporation. Quantification of the frequencies of KI67 + versus KI67 - cells found in each cell cycle phase.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
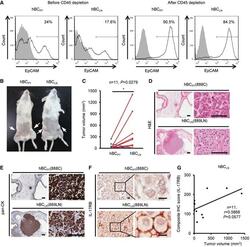
- Figure 8 Elevated IL-17RB expression in cancer cells derived from tumor-draining lymph nodes in breast cancer patients correlates with aggressive growth nature A Freshly collected human breast cancer (hBC) specimens (PT, primary tumors; LN, paired LN metastasis from the same breast cancer patient) were digested with collagenase-containing buffer overnight. Depletion of hematopoietic cells by CD45 Dynabeads (Invitrogen) enriched the EpCAM + cells for following xenografts. Percentages of EpCAM + cells in human primary breast cancer cells were confirmed by FACS analysis (left two panels: before enrichment; right two panels: after enrichment). All of the histograms were shown that they were EpCAM expressing (black line), compared to isotype control (gray filled). B Tumorigenesis assay was determined in NOD/SCID/IL2Rgamma mice orthotopically injected with hBC PT and hBC LN cells. Representative data on day 180 were shown. White arrow: Orthotopic injection site. C Dot plot illustrated the tumor volumes of hBC PT and hBC LN cell-injected NOD/SCID/IL2Rgamma mice on day 180 ( n = 11). All tumors were confirmed by pan-keratin IHC staining of fat pad sections. D, E H&E stainings and IHC stainings of pan-keratin were shown in the representative cases of fat pad sections from (B). Scale bar: 1 mm (left panels), 100 mum (right panels). F IHC stainings of IL-17RB were shown in the representative cases of fat pad sections from (B). Scale bar: 10 mum. G The correlatio
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
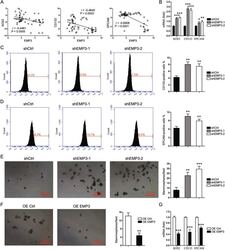
- Fig. 4 EMP3 interferes with stem-like properties. A CCLE database was interrogated for EMP3, SOX2, CD133, and EPCAM expression. Correlation between two genes in a total of 57 breast cancer cell lines was analyzed by Pearson statistics. B SOX2, CD133, and EPCAM expression in MDA-MB-231 cells were detected by real-time PCR. Mean +- SD, n = 3. * P < 0.05, ** P < 0.01, *** P < 0.001, vs. shCtrl. C Fractions of CD133-positive MDA-MB-231 cells were detected by flow cytometry. Mean +- SD, n = 3. ** P < 0.01, vs. shCtrl. D Fractions of EPCAM-positive MDA-MB-231 cells were detected by flow cytometry. Mean +- SD, n = 3. ** P < 0.01, vs. shCtrl. E Mammosphere formation of MDA-MB-231 cells was shown. Mean +- SD, n = 3. ** P < 0.01, *** P < 0.001, vs. shCtrl. F Mammosphere formation of MCF7 cells was shown. Mean +- SD, n = 3. ** P < 0.01, vs. OE Ctrl. G SOX2, CD133, and EPCAM expression in MCF7 cells were detected by real-time PCR. Mean +- SD, n = 3. *** P < 0.001, vs. OE Ctrl.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
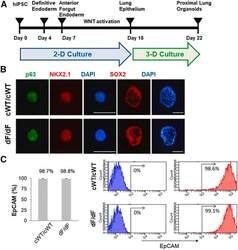
- Figure 1 Generation of proximal lung organoids from cystic fibrosis patient iPSCs (A) Schematic representation of lung organoid differentiation protocol from human iPSCs (hiPSCs). (B) Representative immunofluorescence staining for p63 (green), NKX2.1 (red), SOX2 (red), and nuclear protein expression with DNA stain (DAPI; blue) in HLOs on day 20 (p63 and NKX2.1) and day 22 (SOX2). Scale bars represent 100 mum. (C) Flow cytometry analysis of EpCAM expression in organoid cells on day 23. Left: summary percentage of EpCAM-positive cells. Three independent HLO plates were used for the experiment. Data are presented as mean +- standard error of the mean (SEM). Right: a representative flow cytometry result. The x axis indicates the count of cells, and the y axis indicates EpCAM signal level.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
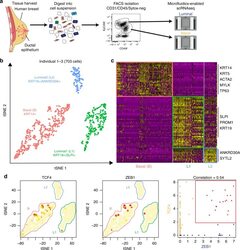
- Fig. 1 Identification of three major epithelial cell types and their markers using scRNAseq. a Overview of scRNAseq approach using primary human breast tissue samples that were processed into single cell suspension, followed by FACS isolation of basal (CD49f-hi, EPCAM+) and luminal (CD49f+, EPCAM-hi), and scRNAseq analysis using the microfluidics-enabled scRNAseq. b Combined tSNE projection of cells from all three microfluidics-enabled scRNAseq datasets. The major basal cluster is highlighted in red; Luminal1 (L1) in green; Luminal2 (L2) in blue. c Heatmap displaying the scaled expression patterns of top marker genes within each cell type with selected marker genes highlighted; yellow indicating high expression of a particular gene, and purple indicating low expression. d Feature plots showing the scaled expression of TCF4 and ZEB1 marking a subpopulation of basal cells and gene plot showing co-expression of TCF4 and ZEB1 in the same cells. See Supplementary Fig. 1 capture site imaging, gene detection, individual principal component analysis, tSNE plot colored by individual-derived cells and feature plots of cell type-specific markers
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
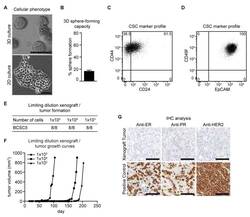
- Figure 1 Characterization of BCSC5 in vitro and in vivo. ( A ) Representative pictures of BCSC5 cultured in 3D and 2D conditions, scale bar 100 mum. ( B ) Sphere-forming capacity of BCSC5 cells in an anchorage-independent growth assay ( n = 3). Data represent means + SEM. ( C , D ) Expression patterns of CD24 and CD44 ( C ) as well as EpCAM and CD49f ( D ) in BCSC5 cells analyzed by flow cytometry. ( E ) Tumor formation in limiting dilution xenografts of BCSC5. ( F ) Representative growth curves for limiting dilution assay of BCSC5 xenografts in immunocompromised NOD/SCID mice. ( G ) Immunohistochemical (IHC) analysis of ER, PR, and HER2 on sections of the BCSC5 xenograft tumors, scale bar 100 mum.
 Explore
Explore Validate
Validate Learn
Learn