13-3600
antibody from Invitrogen Antibodies
Targeting: STAT5A
MGF, STAT5
 Western blot
Western blot ELISA
ELISA Immunocytochemistry
Immunocytochemistry Immunoprecipitation
Immunoprecipitation Immunohistochemistry
Immunohistochemistry Gel shift
Gel shift Other assay
Other assayAntibody data
- Antibody Data
- Antigen structure
- References [21]
- Comments [0]
- Validations
- Immunocytochemistry [2]
- Immunohistochemistry [2]
- Other assay [13]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 13-3600 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- STAT5 alpha Monoclonal Antibody (ST5a-2H2)
- Antibody type
- Monoclonal
- Antigen
- Synthetic peptide
- Description
- This antibody recognizes the STAT5a protein from a variety of species including human, mouse, and rat and it reacts specifically with the STAT5a protein and does not exhibit any cross-reactivity with the highly related STAT5b protein. Cross-reactivity with other endogenous STAT isoforms has not been observed. Reactivity has been confirmed by western blotting using cell lysates derived from human A431 cells (+/- EGF treatment), NIH 3T3 cells, MCF-7 cells, and HeLa cells.
- Reactivity
- Human, Mouse, Rat
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- ST5a-2H2
- Vial size
- 100 μg
- Concentration
- 0.5 mg/mL
- Storage
- -20°C
Submitted references Drug target validation in primary human natural killer cells using CRISPR RNP.
Prospects for Clinical Development of Stat5 Inhibitor IST5-002: High Transcriptomic Specificity in Prostate Cancer and Low Toxicity In Vivo.
Deficiency of core fucosylation activates cellular signaling dependent on FLT3 expression in a Ba/F3 cell system.
Rapid Inflammation in Mice Lacking Both SOCS1 and SOCS3 in Hematopoietic Cells.
Identification of STAT3 and STAT5 proteins in the rat suprachiasmatic nucleus and the Day/Night difference in astrocytic STAT3 phosphorylation in response to lipopolysaccharide.
DNA methylation and transcription in a distal region upstream from the bovine AlphaS1 casein gene after once or twice daily milking.
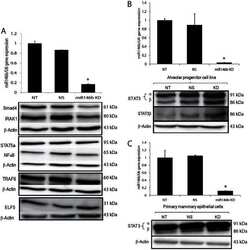
A novel role of microRNA146b in promoting mammary alveolar progenitor cell maintenance.
Preoperative irradiation for the prevention of heterotopic ossification induces local inflammation in humans.
Rac1 signalling modulates a STAT5/BCL-6 transcriptional switch on cell-cycle-associated target gene promoters.
A humanized pattern of aromatase expression is associated with mammary hyperplasia in mice.
Prolactin signalling in the mouse hypothalamus is primarily mediated by signal transducer and activator of transcription factor 5b but not 5a.
Integration of prolactin and glucocorticoid signaling at the beta-casein promoter and enhancer by ordered recruitment of specific transcription factors and chromatin modifiers.
Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility.
The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5.
Hormone-induced modifications of the chromatin structure surrounding upstream regulatory regions conserved between the mouse and rabbit whey acidic protein genes.
Suppressor of cytokine signaling-1 regulates signaling in response to interleukin-2 and other gamma c-dependent cytokines in peripheral T cells.
STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPbeta.
Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation.
Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation.
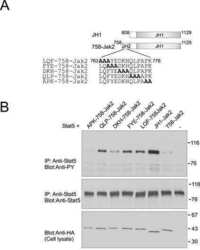
Autoinhibition of Jak2 tyrosine kinase is dependent on specific regions in its pseudokinase domain.
Cooperation among Stat1, glucocorticoid receptor, and PU.1 in transcriptional activation of the high-affinity Fc gamma receptor I in monocytes.
Rautela J, Surgenor E, Huntington ND
Journal of leukocyte biology 2020 Oct;108(4):1397-1408
Journal of leukocyte biology 2020 Oct;108(4):1397-1408
Prospects for Clinical Development of Stat5 Inhibitor IST5-002: High Transcriptomic Specificity in Prostate Cancer and Low Toxicity In Vivo.
Maranto C, Udhane V, Jia J, Verma R, Müller-Newen G, LaViolette PS, Pereckas M, Sabharwal L, Terhune S, Pattabiraman N, Njar VCO, Imig JD, Wang L, Nevalainen MT
Cancers 2020 Nov 18;12(11)
Cancers 2020 Nov 18;12(11)
Deficiency of core fucosylation activates cellular signaling dependent on FLT3 expression in a Ba/F3 cell system.
Duan C, Fukuda T, Isaji T, Qi F, Yang J, Wang Y, Takahashi S, Gu J
FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2020 Feb;34(2):3239-3252
FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2020 Feb;34(2):3239-3252
Rapid Inflammation in Mice Lacking Both SOCS1 and SOCS3 in Hematopoietic Cells.
Ushiki T, Huntington ND, Glaser SP, Kiu H, Georgiou A, Zhang JG, Metcalf D, Nicola NA, Roberts AW, Alexander WS
PloS one 2016;11(9):e0162111
PloS one 2016;11(9):e0162111
Identification of STAT3 and STAT5 proteins in the rat suprachiasmatic nucleus and the Day/Night difference in astrocytic STAT3 phosphorylation in response to lipopolysaccharide.
Moravcová S, Červená K, Pačesová D, Bendová Z
Journal of neuroscience research 2016 Jan;94(1):99-108
Journal of neuroscience research 2016 Jan;94(1):99-108
DNA methylation and transcription in a distal region upstream from the bovine AlphaS1 casein gene after once or twice daily milking.
Nguyen M, Boutinaud M, Pétridou B, Gabory A, Pannetier M, Chat S, Bouet S, Jouneau L, Jaffrezic F, Laloë D, Klopp C, Brun N, Kress C, Jammes H, Charlier M, Devinoy E
PloS one 2014;9(11):e111556
PloS one 2014;9(11):e111556
A novel role of microRNA146b in promoting mammary alveolar progenitor cell maintenance.
Elsarraj HS, Hong Y, Valdez K, Carletti M, Salah SM, Raimo M, Taverna D, Prochasson P, Bharadwaj U, Tweardy DJ, Christenson LK, Behbod F
Journal of cell science 2013 Jun 1;126(Pt 11):2446-58
Journal of cell science 2013 Jun 1;126(Pt 11):2446-58
Preoperative irradiation for the prevention of heterotopic ossification induces local inflammation in humans.
Hoff P, Rakow A, Gaber T, Hahne M, Sentürk U, Strehl C, Fangradt M, Schmidt-Bleek K, Huscher D, Winkler T, Matziolis D, Matziolis G, Badakhshi H, Burmester GR, Duda GN, Perka C, Buttgereit F
Bone 2013 Jul;55(1):93-101
Bone 2013 Jul;55(1):93-101
Rac1 signalling modulates a STAT5/BCL-6 transcriptional switch on cell-cycle-associated target gene promoters.
Barros P, Lam EW, Jordan P, Matos P
Nucleic acids research 2012 Sep;40(16):7776-87
Nucleic acids research 2012 Sep;40(16):7776-87
A humanized pattern of aromatase expression is associated with mammary hyperplasia in mice.
Zhao H, Pearson EK, Brooks DC, Coon JS 5th, Chen D, Demura M, Zhang M, Clevenger CV, Xu X, Veenstra TD, Chatterton RT, DeMayo FJ, Bulun SE
Endocrinology 2012 Jun;153(6):2701-13
Endocrinology 2012 Jun;153(6):2701-13
Prolactin signalling in the mouse hypothalamus is primarily mediated by signal transducer and activator of transcription factor 5b but not 5a.
Yip SH, Eguchi R, Grattan DR, Bunn SJ
Journal of neuroendocrinology 2012 Dec;24(12):1484-91
Journal of neuroendocrinology 2012 Dec;24(12):1484-91
Integration of prolactin and glucocorticoid signaling at the beta-casein promoter and enhancer by ordered recruitment of specific transcription factors and chromatin modifiers.
Kabotyanski EB, Huetter M, Xian W, Rijnkels M, Rosen JM
Molecular endocrinology (Baltimore, Md.) 2006 Oct;20(10):2355-68
Molecular endocrinology (Baltimore, Md.) 2006 Oct;20(10):2355-68
Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility.
Silver DL, Naora H, Liu J, Cheng W, Montell DJ
Cancer research 2004 May 15;64(10):3550-8
Cancer research 2004 May 15;64(10):3550-8
The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5.
Gewinner C, Hart G, Zachara N, Cole R, Beisenherz-Huss C, Groner B
The Journal of biological chemistry 2004 Jan 30;279(5):3563-72
The Journal of biological chemistry 2004 Jan 30;279(5):3563-72
Hormone-induced modifications of the chromatin structure surrounding upstream regulatory regions conserved between the mouse and rabbit whey acidic protein genes.
Millot B, Montoliu L, Fontaine ML, Mata T, Devinoy E
The Biochemical journal 2003 May 15;372(Pt 1):41-52
The Biochemical journal 2003 May 15;372(Pt 1):41-52
Suppressor of cytokine signaling-1 regulates signaling in response to interleukin-2 and other gamma c-dependent cytokines in peripheral T cells.
Cornish AL, Chong MM, Davey GM, Darwiche R, Nicola NA, Hilton DJ, Kay TW, Starr R, Alexander WS
The Journal of biological chemistry 2003 Jun 20;278(25):22755-61
The Journal of biological chemistry 2003 Jun 20;278(25):22755-61
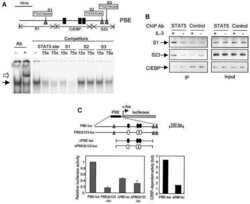
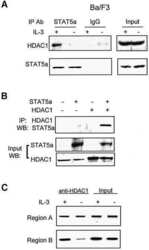
STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPbeta.
Xu M, Nie L, Kim SH, Sun XH
The EMBO journal 2003 Feb 17;22(4):893-904
The EMBO journal 2003 Feb 17;22(4):893-904
Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation.
Chong MM, Cornish AL, Darwiche R, Stanley EG, Purton JF, Godfrey DI, Hilton DJ, Starr R, Alexander WS, Kay TW
Immunity 2003 Apr;18(4):475-87
Immunity 2003 Apr;18(4):475-87
Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation.
Chong MM, Cornish AL, Darwiche R, Stanley EG, Purton JF, Godfrey DI, Hilton DJ, Starr R, Alexander WS, Kay TW
Immunity 2003 Apr;18(4):475-87
Immunity 2003 Apr;18(4):475-87
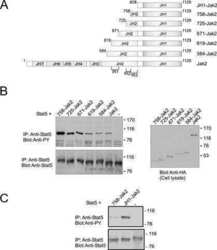
Autoinhibition of Jak2 tyrosine kinase is dependent on specific regions in its pseudokinase domain.
Saharinen P, Vihinen M, Silvennoinen O
Molecular biology of the cell 2003 Apr;14(4):1448-59
Molecular biology of the cell 2003 Apr;14(4):1448-59
Cooperation among Stat1, glucocorticoid receptor, and PU.1 in transcriptional activation of the high-affinity Fc gamma receptor I in monocytes.
Aittomäki S, Pesu M, Groner B, Jänne OA, Palvimo JJ, Silvennoinen O
Journal of immunology (Baltimore, Md. : 1950) 2000 Jun 1;164(11):5689-97
Journal of immunology (Baltimore, Md. : 1950) 2000 Jun 1;164(11):5689-97
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
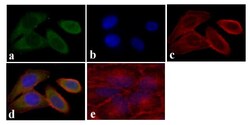
- Experimental details
- Immunofluorescent analysis of STAT5 alpha was done on 70% confluent log phase HeLa cells. The cells were fixed with 4% paraformaldehyde for 15 minutes, permeabilized with 0.25% Triton™ X-100 for 10 minutes, and blocked with 5% BSA for 1 hour at room temperature. The cells were labeled with STAT5 alpha Mouse Monoclonal Antibody (13-3600) at 1:250 dilution in 0.1% BSA and incubated for 3 hours at room temperature and then labeled with Alexa Fluor 488 Rabbit Anti-Mouse IgG Secondary Antibody (Product # A-11059) at a dilution of 1:400 for 30 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (Product # S36938). F-actin (Panel c: red) was stained with Alexa Fluor 594 Phalloidin (Product # A12381). Panel d is a merged image showing cytoplasmic localization. Panel e shows no primary antibody control. The images were captured at 20X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
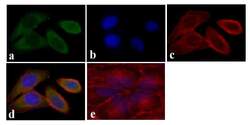
- Experimental details
- Immunofluorescent analysis of STAT5 alpha was done on 70% confluent log phase HeLa cells. The cells were fixed with 4% paraformaldehyde for 15 minutes, permeabilized with 0.25% Triton™ X-100 for 10 minutes, and blocked with 5% BSA for 1 hour at room temperature. The cells were labeled with STAT5 alpha Mouse Monoclonal Antibody (13-3600) at 1:250 dilution in 0.1% BSA and incubated for 3 hours at room temperature and then labeled with Alexa Fluor 488 Rabbit Anti-Mouse IgG Secondary Antibody (Product # A-11059) at a dilution of 1:400 for 30 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (Product # S36938). F-actin (Panel c: red) was stained with Alexa Fluor 594 Phalloidin (Product # A12381). Panel d is a merged image showing cytoplasmic localization. Panel e shows no primary antibody control. The images were captured at 20X magnification.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
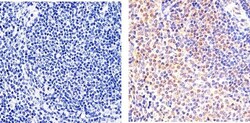
- Experimental details
- Immunohistochemistry analysis of STAT5 alpha showing staining in the cytoplasm and nucleus of paraffin-embedded mouse spleen tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a STAT5 alpha monoclonal antibody (Product # 13-3600) diluted in 3% BSA-PBS at a dilution of 1:20 overnight at 4°C in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
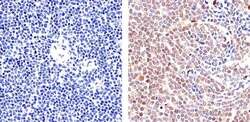
- Experimental details
- Immunohistochemistry analysis of STAT5 alpha showing staining in the cytoplasm and nucleus of paraffin-embedded human T cell lymphoma (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a STAT5 alpha monoclonal antibody (Product # 13-3600) diluted in 3% BSA-PBS at a dilution of 1:20 overnight at 4°C in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
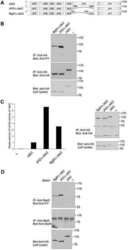
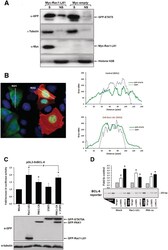
- Figure 2. Rac1 signalling switches promoter occupancy from BCL-6 to STAT5. ( A ) DLD-1 cells were transfected with Myc-Rac1-Q61L or control empty vector and 24 h later analyzed by Western blot for the subcellular distribution of STAT5 between a soluble (S) and a chromatin-bound non-soluble (NS) fraction (detection of alpha-tubulin and histone 2B served as controls). Note that active Rac1 promotes retention of STAT5 in the non-soluble chromatin fraction. ( B ) Subcellular localization of STAT5 determined by confocal fluorescence microscopy in DLD-1 cells co-transfected with DsRed-Rac1-Q61L and GFP-STAT5A. The overlay image of the DAPI, GFP and DsRed channels is shown. A microscopic field was chosen that contained side by side untransfected cells (blue nuclei), cells that transfected only with GFP-STAT5 (green cells) and cells that co-transfected with both GFP-STAT5 and DsRed-Rac1-Q61L (red cells). Note the nuclear STAT5 signal in Rac1-expressing red cells. In addition, two plots are given showing the DAPI and GFP signal intensities measured along the indicated regions of interest (ROI, white lines). The signal intensity of GFP did not increase across the nuclear DAPI region when cells expressed only GFP-STAT5 (green cells, ROI 1), whereas nuclear GFP signal clearly increased when cells co-expressed active Rac1 (red cells, ROI 2), confirming nuclear translocation of GFP-STAT5. ( C ) DLD-1 cells were co-transfected with the reporter and the indicated expression vectors, as descr
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
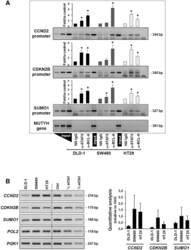
- Figure 5. Regulation of expression of the CCND2, CDKN2B and SUMO1 genes. ( A ) Promoter occupancies with BCL-6 and STAT5 at the CCND2, CDKN2B and SUMO1 gene promoters was determined by ChIP with the indicated antibodies using lysates of the three indicated cell lines (see legend to Figure 2 D for details). A representative semi-qPCR of the precipitated promoter fragments and a graphical representation of the respective band intensities, * P < 0.05, are shown. A control genomic fragment from the MUTYH gene was amplified to confirm the specificity of the precipitated target gene promoters. Note that BCL-6 binds predominantly in the PAK1-lacking SW480 cells and whereas a switch to STAT5 occurs in HT29 cells with active Rac1/PAK1 signalling. ( B ) Gene expression data corresponding to the ChIP analysis of the three genes in the three cell lines. Left panel shows representative semi-quantitative reverse transcriptase-PCRs (RT-PCRs), whereas graph at the right shows the result of qPCR analysis of cDNAs collected from the three cell lines at three different splitting times. Genes encoding RNA polymerase II (pol 2) and the glycolytic enzyme phosphoglycerate kinase (PGK1) were amplified as control housekeeping genes and a pool of cDNAs mixed at equal parts from the three cell lines was used as reference for qPCR. Serial dilutions served to assure semi-quantitative conditions in the conventional RT-PCR reactions.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
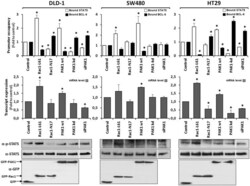
- Figure 6. Rac1 signalling controls target gene expression by inverting promoter occupancy with either BCL-6 or STAT5. The representative analysis of the SUMO1 gene is shown in the indicated three colorectal cell lines following their transfection with constructs that either activate or inhibit Rac1 signalling. Top panels show the graphical display of promoter occupancy by ChIP using either anti-BCL-6 (black columns) or STAT5 (white columns) and middle panels the respective gene expression levels (grey columns) (see legend to Figure 5 for further details), * P < 0.05. Bottom panels show Western blot analysis of the cell lysates demonstrating the expression levels of the transfected GFP, GFP-Rac1 or GFP-PAK1 constructs as well as the resulting phosphorylation status of endogenous STAT5. Note that in SW480, which lack endogenous PAK1, depletion of endogenous PAK1 by siRNAs transfection (documented in Fig. S2B ) or expression of dominant negative PAK1 has no effect on promoter-bound BCL-6, whereas re-expression of PAK1 leads to loss of BCL-6 from the SUMO1 promoter and an increase in gene expression. In the other two cell lines, inhibition of Rac1 or PAK1 are clearly correlated with more BCL-6 bound and less gene expression while activation of Rac1 or PAK1 promoted STAT5 binding to the promoters and increased transcription.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
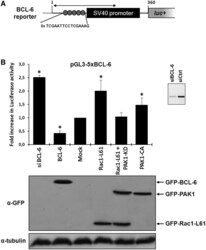
- Figure 1. Rac1 signalling promotes transcription by repressing BCL-6 and stimulating STAT5. ( A ) Schematic representation of the transcriptional luciferase reporter vector under the control of five consensus BCL-6 binding motifs. ( B ) DLD-1 cells were co-transfected with the reporter vector and the indicated expression vectors or siRNAs. Cells were lysed 24 h later and luciferase activity was measured and graphically displayed, * P < 0.05. The Western blot below the graph shows the levels of transfected GFP-tagged BCL-6, Rac1-L61, PAK1 kinase dead (kd) or PAK1 constitutively active (ca) mutants. Detection of endogenous alpha-tubulin served as loading control. The small insert beside the graph shows a Western blot of endogenous BCL-6 to document the efficiency of its siRNA-mediated depletion.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
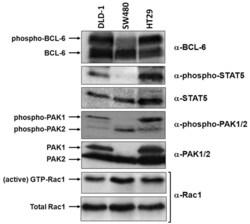
- Figure 3. Correlation of Rac1 signalling and activation of PAK1, BCL-6 or STAT5 in different cell lines. Equivalent lysate quantities of DLD-1, SW480 and HT29 colorectal cells were separated by gel electrophoresis and analysed by Western blot using the indicated antibodies to compare protein levels. The active Rac1 fraction was obtained by CRIB-pull down assays, as described ( 6 ).
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
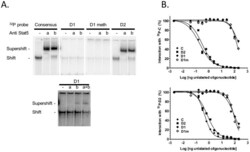
- Figure 6 Binding activity of two potential STAT5 binding sites located within the CSN1S1 distal upstream region. (A) Representative EMSA. Mammary nuclear extract (22ug) from rabbits in early lactation were incubated with the indicated labelled 32 P probes in the presence or absence of STAT5a or STAT5b antibodies (1 or 0.5 ug, respectively). Complexes were analysed on non denaturating 5% acrylamide gels in 0.25xTBE. The lower panel is an overexposed image of the upper one to show the weak interaction between D1 and nuclear extracts and the supershifts with STAT5 antibodies. (B) Competition assays with increasing amounts of unlabelled C, D2, D1 and D1m using 32 P-C or 32 P-D2, are indicated in each graph. Results are expressed as a percentage of the radioactivity bound in the absence of unlabelled oligonucleotides and plotted as a function of the log (ng oligonucleotide) in the reaction.
 Explore
Explore Validate
Validate Learn
Learn