Antibody data
- Antibody Data
- Antigen structure
- References [42]
- Comments [0]
- Validations
- Flow cytometry [1]
- Other assay [45]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 71-8700 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- Cullin 1 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Synthetic peptide
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μg
- Concentration
- 0.25 mg/mL
- Storage
- -20°C
Submitted references Entamoeba histolytica activation of caspase-1 degrades cullin that attenuates NF-κB dependent signaling from macrophages.
FBXO44 promotes DNA replication-coupled repetitive element silencing in cancer cells.
A class of viral inducer of degradation of the necroptosis adaptor RIPK3 regulates virus-induced inflammation.
FBXW7 Triggers Degradation of KMT2D to Favor Growth of Diffuse Large B-cell Lymphoma Cells.
Overexpression of IFIT2 inhibits the proliferation of chronic myeloid leukemia cells by regulating the BCR‑ABL/AKT/mTOR pathway.
FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2Fe2S] Cluster.
The Protein Neddylation Inhibitor MLN4924 Suppresses Patient-Derived Glioblastoma Cells via Inhibition of ERK and AKT Signaling.
Unanchored tri-NEDD8 inhibits PARP-1 to protect from oxidative stress-induced cell death.
The F-Box Domain-Dependent Activity of EMI1 Regulates PARPi Sensitivity in Triple-Negative Breast Cancers.
Partial proteasomal degradation of Lola triggers the male-to-female switch of a dimorphic courtship circuit.
Loss of KLHL6 promotes diffuse large B-cell lymphoma growth and survival by stabilizing the mRNA decay factor roquin2.
PTEN counteracts FBXL2 to promote IP3R3- and Ca(2+)-mediated apoptosis limiting tumour growth.
Targeted inhibition of the COP9 signalosome for treatment of cancer.
Characterization of the mammalian family of DCN-type NEDD8 E3 ligases.
COP9-Signalosome deneddylase activity is enhanced by simultaneous neddylation: insights into the regulation of an enzymatic protein complex.
CSN6 drives carcinogenesis by positively regulating Myc stability.
Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins.
HERC2 targets the iron regulator FBXL5 for degradation and modulates iron metabolism.
Human papillomavirus type 16 E7 oncoprotein inhibits the anaphase promoting complex/cyclosome activity by dysregulating EMI1 expression in mitosis.
Selective enhancing effect of early mitotic inhibitor 1 (Emi1) depletion on the sensitivity of doxorubicin or X-ray treatment in human cancer cells.
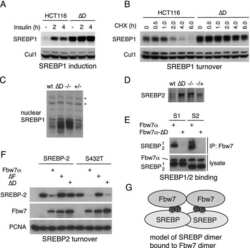
Fbw7 dimerization determines the specificity and robustness of substrate degradation.
β-TrCP-mediated IRAK1 degradation releases TAK1-TRAF6 from the membrane to the cytosol for TAK1-dependent NF-κB activation.
Cullin-3 regulates late endosome maturation.
Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system.
UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1α accumulation.
NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway.
SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation.
The human COP9 signalosome protects ubiquitin-conjugating enzyme 3 (UBC3/Cdc34) from beta-transducin repeat-containing protein (betaTrCP)-mediated degradation.
Regulation of postsynaptic RapGAP SPAR by Polo-like kinase 2 and the SCFbeta-TRCP ubiquitin ligase in hippocampal neurons.
Diversity in tissue expression, substrate binding, and SCF complex formation for a lectin family of ubiquitin ligases.
TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex.
Selective cochlear degeneration in mice lacking the F-box protein, Fbx2, a glycoprotein-specific ubiquitin ligase subunit.
Substrate-mediated regulation of cullin neddylation.
The COP9 signalosome regulates Skp2 levels and proliferation of human cells.
A novel route for F-box protein-mediated ubiquitination links CHIP to glycoprotein quality control.
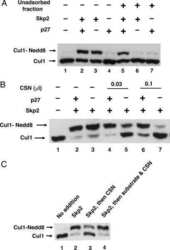
Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate.
SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein.
Multiple functions of Jab1 are required for early embryonic development and growth potential in mice.
Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage.
The F-box protein SKP2 mediates androgen control of p27 stability in LNCaP human prostate cancer cells.
The F-box protein SKP2 mediates androgen control of p27 stability in LNCaP human prostate cancer cells.
A dominant-negative UBC12 mutant sequesters NEDD8 and inhibits NEDD8 conjugation in vivo.
Chadha A, Moreau F, Wang S, Dufour A, Chadee K
PLoS pathogens 2021 Sep;17(9):e1009936
PLoS pathogens 2021 Sep;17(9):e1009936
FBXO44 promotes DNA replication-coupled repetitive element silencing in cancer cells.
Shen JZ, Qiu Z, Wu Q, Finlay D, Garcia G, Sun D, Rantala J, Barshop W, Hope JL, Gimple RC, Sangfelt O, Bradley LM, Wohlschlegel J, Rich JN, Spruck C
Cell 2021 Jan 21;184(2):352-369.e23
Cell 2021 Jan 21;184(2):352-369.e23
A class of viral inducer of degradation of the necroptosis adaptor RIPK3 regulates virus-induced inflammation.
Liu Z, Nailwal H, Rector J, Rahman MM, Sam R, McFadden G, Chan FK
Immunity 2021 Feb 9;54(2):247-258.e7
Immunity 2021 Feb 9;54(2):247-258.e7
FBXW7 Triggers Degradation of KMT2D to Favor Growth of Diffuse Large B-cell Lymphoma Cells.
Saffie R, Zhou N, Rolland D, Önder Ö, Basrur V, Campbell S, Wellen KE, Elenitoba-Johnson KSJ, Capell BC, Busino L
Cancer research 2020 Jun 15;80(12):2498-2511
Cancer research 2020 Jun 15;80(12):2498-2511
Overexpression of IFIT2 inhibits the proliferation of chronic myeloid leukemia cells by regulating the BCR‑ABL/AKT/mTOR pathway.
Zhang Z, Li N, Liu S, Jiang M, Wan J, Zhang Y, Wan L, Xie C, Le A
International journal of molecular medicine 2020 Apr;45(4):1187-1194
International journal of molecular medicine 2020 Apr;45(4):1187-1194
FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2Fe2S] Cluster.
Wang H, Shi H, Rajan M, Canarie ER, Hong S, Simoneschi D, Pagano M, Bush MF, Stoll S, Leibold EA, Zheng N
Molecular cell 2020 Apr 2;78(1):31-41.e5
Molecular cell 2020 Apr 2;78(1):31-41.e5
The Protein Neddylation Inhibitor MLN4924 Suppresses Patient-Derived Glioblastoma Cells via Inhibition of ERK and AKT Signaling.
Han S, Shin H, Oh JW, Oh YJ, Her NG, Nam DH
Cancers 2019 Nov 22;11(12)
Cancers 2019 Nov 22;11(12)
Unanchored tri-NEDD8 inhibits PARP-1 to protect from oxidative stress-induced cell death.
Keuss MJ, Hjerpe R, Hsia O, Gourlay R, Burchmore R, Trost M, Kurz T
The EMBO journal 2019 Mar 15;38(6)
The EMBO journal 2019 Mar 15;38(6)
The F-Box Domain-Dependent Activity of EMI1 Regulates PARPi Sensitivity in Triple-Negative Breast Cancers.
Marzio A, Puccini J, Kwon Y, Maverakis NK, Arbini A, Sung P, Bar-Sagi D, Pagano M
Molecular cell 2019 Jan 17;73(2):224-237.e6
Molecular cell 2019 Jan 17;73(2):224-237.e6
Partial proteasomal degradation of Lola triggers the male-to-female switch of a dimorphic courtship circuit.
Sato K, Ito H, Yokoyama A, Toba G, Yamamoto D
Nature communications 2019 Jan 11;10(1):166
Nature communications 2019 Jan 11;10(1):166
Loss of KLHL6 promotes diffuse large B-cell lymphoma growth and survival by stabilizing the mRNA decay factor roquin2.
Choi J, Lee K, Ingvarsdottir K, Bonasio R, Saraf A, Florens L, Washburn MP, Tadros S, Green MR, Busino L
Nature cell biology 2018 May;20(5):586-596
Nature cell biology 2018 May;20(5):586-596
PTEN counteracts FBXL2 to promote IP3R3- and Ca(2+)-mediated apoptosis limiting tumour growth.
Kuchay S, Giorgi C, Simoneschi D, Pagan J, Missiroli S, Saraf A, Florens L, Washburn MP, Collazo-Lorduy A, Castillo-Martin M, Cordon-Cardo C, Sebti SM, Pinton P, Pagano M
Nature 2017 Jun 22;546(7659):554-558
Nature 2017 Jun 22;546(7659):554-558
Targeted inhibition of the COP9 signalosome for treatment of cancer.
Schlierf A, Altmann E, Quancard J, Jefferson AB, Assenberg R, Renatus M, Jones M, Hassiepen U, Schaefer M, Kiffe M, Weiss A, Wiesmann C, Sedrani R, Eder J, Martoglio B
Nature communications 2016 Oct 24;7:13166
Nature communications 2016 Oct 24;7:13166
Characterization of the mammalian family of DCN-type NEDD8 E3 ligases.
Keuss MJ, Thomas Y, Mcarthur R, Wood NT, Knebel A, Kurz T
Journal of cell science 2016 Apr 1;129(7):1441-54
Journal of cell science 2016 Apr 1;129(7):1441-54
COP9-Signalosome deneddylase activity is enhanced by simultaneous neddylation: insights into the regulation of an enzymatic protein complex.
Bornstein G, Grossman C
Cell division 2015;10:5
Cell division 2015;10:5
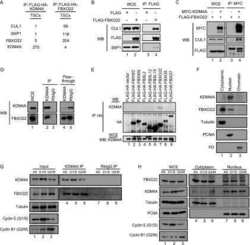
CSN6 drives carcinogenesis by positively regulating Myc stability.
Chen J, Shin JH, Zhao R, Phan L, Wang H, Xue Y, Post SM, Ho Choi H, Chen JS, Wang E, Zhou Z, Tseng C, Gully C, Velazquez-Torres G, Fuentes-Mattei E, Yeung G, Qiao Y, Chou PC, Su CH, Hsieh YC, Hsu SL, Ohshiro K, Shaikenov T, Wang H, Yeung SC, Lee MH
Nature communications 2014 Nov 14;5:5384
Nature communications 2014 Nov 14;5:5384
Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins.
Gong Y, Zack TI, Morris LG, Lin K, Hukkelhoven E, Raheja R, Tan IL, Turcan S, Veeriah S, Meng S, Viale A, Schumacher SE, Palmedo P, Beroukhim R, Chan TA
Nature genetics 2014 Jun;46(6):588-94
Nature genetics 2014 Jun;46(6):588-94
HERC2 targets the iron regulator FBXL5 for degradation and modulates iron metabolism.
Moroishi T, Yamauchi T, Nishiyama M, Nakayama KI
The Journal of biological chemistry 2014 Jun 6;289(23):16430-41
The Journal of biological chemistry 2014 Jun 6;289(23):16430-41
Human papillomavirus type 16 E7 oncoprotein inhibits the anaphase promoting complex/cyclosome activity by dysregulating EMI1 expression in mitosis.
Yu Y, Munger K
Virology 2013 Nov;446(1-2):251-9
Virology 2013 Nov;446(1-2):251-9
Selective enhancing effect of early mitotic inhibitor 1 (Emi1) depletion on the sensitivity of doxorubicin or X-ray treatment in human cancer cells.
Shimizu N, Nakajima NI, Tsunematsu T, Ogawa I, Kawai H, Hirayama R, Fujimori A, Yamada A, Okayasu R, Ishimaru N, Takata T, Kudo Y
The Journal of biological chemistry 2013 Jun 14;288(24):17238-52
The Journal of biological chemistry 2013 Jun 14;288(24):17238-52
Fbw7 dimerization determines the specificity and robustness of substrate degradation.
Welcker M, Larimore EA, Swanger J, Bengoechea-Alonso MT, Grim JE, Ericsson J, Zheng N, Clurman BE
Genes & development 2013 Dec 1;27(23):2531-6
Genes & development 2013 Dec 1;27(23):2531-6
β-TrCP-mediated IRAK1 degradation releases TAK1-TRAF6 from the membrane to the cytosol for TAK1-dependent NF-κB activation.
Cui W, Xiao N, Xiao H, Zhou H, Yu M, Gu J, Li X
Molecular and cellular biology 2012 Oct;32(19):3990-4000
Molecular and cellular biology 2012 Oct;32(19):3990-4000
Cullin-3 regulates late endosome maturation.
Huotari J, Meyer-Schaller N, Hubner M, Stauffer S, Katheder N, Horvath P, Mancini R, Helenius A, Peter M
Proceedings of the National Academy of Sciences of the United States of America 2012 Jan 17;109(3):823-8
Proceedings of the National Academy of Sciences of the United States of America 2012 Jan 17;109(3):823-8
Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system.
Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, Stadtfeld M, Hochedlinger K, Chen EI, Aifantis I
Cell stem cell 2012 Dec 7;11(6):783-98
Cell stem cell 2012 Dec 7;11(6):783-98
UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1α accumulation.
Bandau S, Knebel A, Gage ZO, Wood NT, Alexandru G
BMC biology 2012 Apr 26;10:36
BMC biology 2012 Apr 26;10:36
NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway.
den Besten W, Verma R, Kleiger G, Oania RS, Deshaies RJ
Nature structural & molecular biology 2012 Apr 1;19(5):511-6, S1
Nature structural & molecular biology 2012 Apr 1;19(5):511-6, S1
SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation.
Tan MK, Lim HJ, Harper JW
Molecular and cellular biology 2011 Sep;31(18):3687-99
Molecular and cellular biology 2011 Sep;31(18):3687-99
The human COP9 signalosome protects ubiquitin-conjugating enzyme 3 (UBC3/Cdc34) from beta-transducin repeat-containing protein (betaTrCP)-mediated degradation.
Fernandez-Sanchez ME, Sechet E, Margottin-Goguet F, Rogge L, Bianchi E
The Journal of biological chemistry 2010 Jun 4;285(23):17390-7
The Journal of biological chemistry 2010 Jun 4;285(23):17390-7
Regulation of postsynaptic RapGAP SPAR by Polo-like kinase 2 and the SCFbeta-TRCP ubiquitin ligase in hippocampal neurons.
Ang XL, Seeburg DP, Sheng M, Harper JW
The Journal of biological chemistry 2008 Oct 24;283(43):29424-32
The Journal of biological chemistry 2008 Oct 24;283(43):29424-32
Diversity in tissue expression, substrate binding, and SCF complex formation for a lectin family of ubiquitin ligases.
Glenn KA, Nelson RF, Wen HM, Mallinger AJ, Paulson HL
The Journal of biological chemistry 2008 May 9;283(19):12717-29
The Journal of biological chemistry 2008 May 9;283(19):12717-29
TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex.
Tian Y, Kolb R, Hong JH, Carroll J, Li D, You J, Bronson R, Yaffe MB, Zhou J, Benjamin T
Molecular and cellular biology 2007 Sep;27(18):6383-95
Molecular and cellular biology 2007 Sep;27(18):6383-95
Selective cochlear degeneration in mice lacking the F-box protein, Fbx2, a glycoprotein-specific ubiquitin ligase subunit.
Nelson RF, Glenn KA, Zhang Y, Wen H, Knutson T, Gouvion CM, Robinson BK, Zhou Z, Yang B, Smith RJ, Paulson HL
The Journal of neuroscience : the official journal of the Society for Neuroscience 2007 May 9;27(19):5163-71
The Journal of neuroscience : the official journal of the Society for Neuroscience 2007 May 9;27(19):5163-71
Substrate-mediated regulation of cullin neddylation.
Chew EH, Hagen T
The Journal of biological chemistry 2007 Jun 8;282(23):17032-40
The Journal of biological chemistry 2007 Jun 8;282(23):17032-40
The COP9 signalosome regulates Skp2 levels and proliferation of human cells.
Denti S, Fernandez-Sanchez ME, Rogge L, Bianchi E
The Journal of biological chemistry 2006 Oct 27;281(43):32188-96
The Journal of biological chemistry 2006 Oct 27;281(43):32188-96
A novel route for F-box protein-mediated ubiquitination links CHIP to glycoprotein quality control.
Nelson RF, Glenn KA, Miller VM, Wen H, Paulson HL
The Journal of biological chemistry 2006 Jul 21;281(29):20242-51
The Journal of biological chemistry 2006 Jul 21;281(29):20242-51
Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate.
Bornstein G, Ganoth D, Hershko A
Proceedings of the National Academy of Sciences of the United States of America 2006 Aug 1;103(31):11515-20
Proceedings of the National Academy of Sciences of the United States of America 2006 Aug 1;103(31):11515-20
SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein.
Shirogane T, Jin J, Ang XL, Harper JW
The Journal of biological chemistry 2005 Jul 22;280(29):26863-72
The Journal of biological chemistry 2005 Jul 22;280(29):26863-72
Multiple functions of Jab1 are required for early embryonic development and growth potential in mice.
Tomoda K, Yoneda-Kato N, Fukumoto A, Yamanaka S, Kato JY
The Journal of biological chemistry 2004 Oct 8;279(41):43013-8
The Journal of biological chemistry 2004 Oct 8;279(41):43013-8
Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage.
Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF
Nature 2003 Nov 6;426(6962):87-91
Nature 2003 Nov 6;426(6962):87-91
The F-box protein SKP2 mediates androgen control of p27 stability in LNCaP human prostate cancer cells.
Lu L, Schulz H, Wolf DA
BMC cell biology 2002 Aug 20;3:22
BMC cell biology 2002 Aug 20;3:22
The F-box protein SKP2 mediates androgen control of p27 stability in LNCaP human prostate cancer cells.
Lu L, Schulz H, Wolf DA
BMC cell biology 2002 Aug 20;3:22
BMC cell biology 2002 Aug 20;3:22
A dominant-negative UBC12 mutant sequesters NEDD8 and inhibits NEDD8 conjugation in vivo.
Wada H, Yeh ET, Kamitani T
The Journal of biological chemistry 2000 Jun 2;275(22):17008-15
The Journal of biological chemistry 2000 Jun 2;275(22):17008-15
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
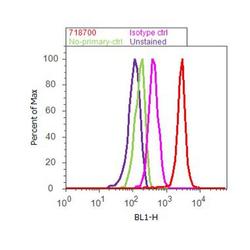
- Experimental details
- Flow cytometry analysis of CUL-1 was done on MDA-MB-231 cells. Cells were fixed with 70% ethanol for 10 minutes, permeabilized with 0.25% Triton™ X-100 for 20 minutes, and blocked with 5% BSA for 30 minutes at room temperature. Cells were labeled with CUL-1 Rabbit Polyclonal Antibody (718700, red histogram) or with rabbit isotype control (pink histogram) at 3-5 ug/million cells in 2.5% BSA. After incubation at room temperature for 2 hours, the cells were labeled with Alexa Fluor® 488 Goat Anti-Rabbit Secondary Antibody (A11008) at a dilution of 1:400 for 30 minutes at room temperature. The representative 10,000 cells were acquired and analyzed for each sample using an Attune® Acoustic Focusing Cytometer. The purple histogram represents unstained control cells and the green histogram represents no-primary-antibody control.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
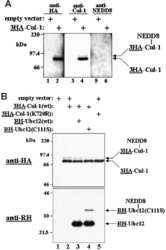
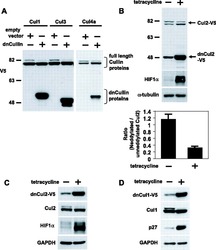
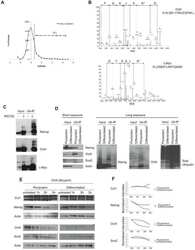
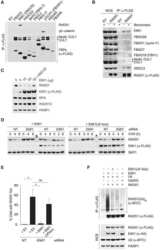
- Fig. 2 ATP-depletion resulted in the inhibition of CSN deneddylase activity. a 10 nM His-6-Cul1/ROC1 were pre-neddylated for 30 min at 30degC. Then 10 mug HeLa cells extract were added at 30degC (time plot: lanes 3 - 6 ); or alternatively, ATP-trap mix was added for 45 min at 30degC, followed by the addition of 10 mug HeLa cells extract at 30degC (time plot: lanes 8 - 11 ). b 3 nM His-6-Cul1/ROC1 was pre-neddylated for 30 min at 30degC. Then 100 nM purified CSN was added at 20degC (time plot: lanes 3 - 6 ); or alternatively, ATP-trap mix was added for 45 min at 30degC, followed by the addition of 100 nM purified CSN at 20degC (time plot: lanes 8 - 11 ). c 10 nM His-6-Cul1/ROC1 was incubated as described in b ; time plots were for 1.5-7.5 min, and not for 5-40 min as in b . Blotting was directed against NEDD8, and not against Cul1, as described elsewhere.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
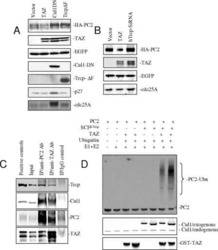
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
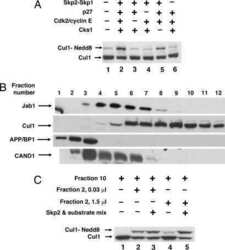
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
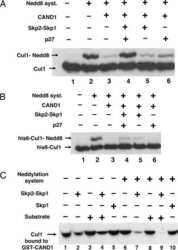
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Fig. 4 Endogenic Cul1 deneddylation is enhanced during simultaneous neddylation. a 20 mug HeLa cells extract were incubated for 30 min at 30degC ( lanes 2 - 7 ) or with the addition of ATP-trap mix prior to the incubation ( lanes 8 - 13 ). Simultaneous ATP-depletion was performed in lanes 3 - 7 after 30 min incubation, on a time plot of 1.5-7.5 min at 30degC. Simultaneous neddylation was performed in lanes 9 - 13 after 30 min, by the addition of AMP-PNP, on a time plot of 1.5-7.5 min at 30degC. b 10 nM His-6-Cul1/ROC1 was neddylated at 30degC on a time plot of 2.5-30 min, without or with the addition of ATP-trap mix ( lanes 2 - 6 and lanes 7 - 11 , respectively). In lane 12 , the neddylation mix was added to the reaction after pre-incubation with ATP-trap mix. c 10 nM His-6-Cul1/ROC1 was neddylated at 30degC using AMP-PNP on a time plot of 2.5-30 min, either directly ( lanes 2 - 6 ) or after pre-incubation with ATP-trap mix ( lanes 7 - 11 ).
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
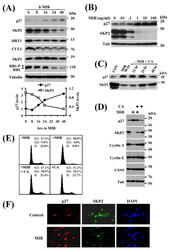
- Figure 2 Effect of MIB on p27, SKP2, and other cell cycle regulators ( A ) LNCaP cells were incubated with 10 ng/ml MIB for the indicated periods. Cell lysate was harvested and expression of the indicated proteins was assessed by immunoblotting with respective antibodies. Tubulin is shown as a loading control. SKP2 and p27 protein levels were quantitated using a demonstration copy of the Totallab software from Nonlinear Dynamics. ( B ) LNCaP cells were incubated with the indicated concentrations of MIB for 72 h, followed by preparation of total cell lysate and immunoblotting with antibodies against p27, SKP2, and tubulin. The effects of androgen on p27 and SKP2 were maximal at 1 ng/ml MIB. ( C ) LNCaP cells were maintained in the presence of 1 ng/ml MIB for 96 h after which the medium was replaced and 750 ng/ml of the antiandrogen cyproterone acetate (CA) was added. Cell lysates were prepared after the indicated times, and p27 and SKP2 levels were assessed by immunoblotting. ( D ) LNCaP cells were incubated with 1 ng/ml MIB and/or 750 ng/ml CA for 72 h and cell lysates were prepared. Expression of the indicated proteins was assessed by immunoblotting. Tubulin levels are shown as loading controls. ( E ) LNCaP cells were incubated with 1 ng/ml MIB and/or 750 ng/ml CA for 72 h and cells were fixed for flow cytometry. Cell cycle profiles and the fraction of cells in each cell cycle phase are shown. ( F ) LNCaP cells were maintained in the absence or presence of 10 ng/ml MIB for 7
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
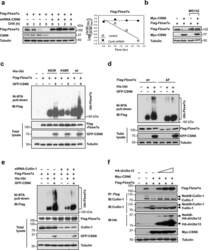
- Figure 3 CSN6 enhances Fbxw7's autoubiquitination and degradation (a) CSN6 increased Fbxw7alpha turnover. 293T cells were infected with lentivirus to express CSN6 shRNA or luciferase control shRNA. The cells were then transfected with Flag-Fbxw7alpha and were then treated with cycloheximide (CHX; 100 ug/ml) for the indicated times. The density of Fbxw7alpha was measured and the integrated optical density (OD) was measured. The turnover of Fbxw7alpha is indicated graphically. (b) CSN6-mediated Fbxw7alpha downregulation was proteasome-dependent. 293T cells were cotransfected with indicated plasmids and were treated with or without 50 ug/ml MG132 for 6 hours. The cell lysates were then immunoblotted with the indicated antibodies. (c) CSN6 enhanced Fbxw7alpha ubiquitination through lysine 48 linkage. 293T cells were cotransfected with Flag-Fbxw7alpha with or without GFP-CSN6 plus His-ubiquitin wild type (wt), K63R mutant, or K48R mutant. Cells were treated with 50 ug/ml MG132 for 6 hours before harvest. The ubiquitinated Fbxw7alpha proteins were pulled down using Ni-NTA-agarose beads and detected with anti-Flag antibody. (d) An F-box domain deletion mutant of Fbxw7alpha was resistant to CSN6-mediated ubiquitination. 293T cells were cotransfected with GFP-CSN6, His-ubiquitin plus Flag-Fbxw7alpha wild-type or F-box domain deletion mutant (DeltaF). Cells were treated with 50 ug/ml MG132 for 6 hours before harvest. The ubiquitinated Fbxw7alpha proteins were pulled down using Ni-
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
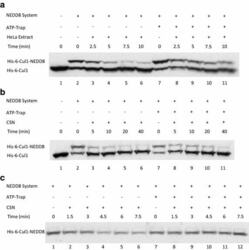
- Fig. 2 ATP-depletion resulted in the inhibition of CSN deneddylase activity. a 10 nM His-6-Cul1/ROC1 were pre-neddylated for 30 min at 30degC. Then 10 mug HeLa cells extract were added at 30degC (time plot: lanes 3 - 6 ); or alternatively, ATP-trap mix was added for 45 min at 30degC, followed by the addition of 10 mug HeLa cells extract at 30degC (time plot: lanes 8 - 11 ). b 3 nM His-6-Cul1/ROC1 was pre-neddylated for 30 min at 30degC. Then 100 nM purified CSN was added at 20degC (time plot: lanes 3 - 6 ); or alternatively, ATP-trap mix was added for 45 min at 30degC, followed by the addition of 100 nM purified CSN at 20degC (time plot: lanes 8 - 11 ). c 10 nM His-6-Cul1/ROC1 was incubated as described in b ; time plots were for 1.5-7.5 min, and not for 5-40 min as in b . Blotting was directed against NEDD8, and not against Cul1, as described elsewhere.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
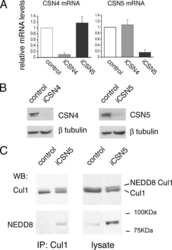
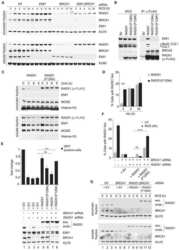
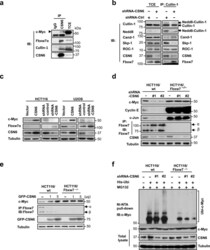
- Figure 2 CSN6 regulates SCF (Skp1/Cullin-1/Fbxw7) complex to stabilize Myc (a) CSN6 associated with Myc, Fbxw7 and Cullin-1. The U2OS cell lysates were immunoprecipitated with anti-CSN6 and immunoblotted with indicated antibodies. (b) CSN6 knockdown led to decreased Cullin-1 neddylation. The U2OS cell lysates were immunoprecipitated with anti-Cullin-1, followed by immunoblotting with indicated antibodies. (c) CSN6 expression was inversely related to Fbxw7 expression. Immunoblot analysis of Myc and Fbxw7alpha in CSN6-overexpressing or CSN6-knockdown cells. Cell lysates were analyzed by indicated antibodies. (d) Knockdown of CSN6 increased the expression of Fbxw7 and reduced Myc. Indicated cells were infected with lentivirus to express one of two independent CSN6 shRNAs (#1, #2) or luciferase control shRNA. Cell lysates were immunoblotted with indicated antibodies. (e) CSN6 reduced steady-state expression of Fbxw7 to upregulate Myc. Indicated cells were transfected with increasing amounts of GFP-CSN6. Cell lysates were immunoblotted with indicated antibodies. Fbxw7alpha and Fbxw7beta isoforms were identified. (h) Knockdown of CSN6 enhanced Myc ubiquitination. Indicated cells were infected with lentivirus to express one of two independent CSN6 shRNAs (#1, #2) or luciferase control shRNA. The cells were then transfected with His-ubiquitin and treated with MG132 for 6 hours before harvest. The ubiquitinated Myc proteins were pulled down using Ni-NTA-agarose beads and detected with
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
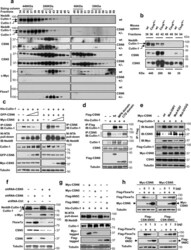
- Figure 4 CSN6 enhances neddylation of Cullin-1 via its MPN domain (a) Neddylation status of Cullin in gel filtration chromatography fractions from spleen extracts from Csn6 +/+ (wt) and Csn6 +/- mice. Extracts of spleen from CSN6 +/+ or Csn6 +/- mice (4 weeks old) were ground and subjected to lysis. Lysates were fractionated by gel filtration chromatography. Fractions were resolved by SDS-PAGE, followed by immunoblotting with indicated antibodies. Molecular size of eluted fraction is indicated above. (b) Non-neddylated Cullin was increased in Csn6 +/- mice. Extracts of spleen from CSN6 +/+ (wt) or Csn6 +/- mice were fractionated by gel filtration chromatography. Representative fractions 36-54 from (a) were resolved by SDS-PAGE, followed by immunoblotting with anti-Cullin-1, anti-CSN6, and anti-CSN5 antibodies. (c) CSN6 competed with CSN5 for binding to Cullin-1 and affected Cullin-1 neddylation. 293T cells were transfected with His-Cullin-1 plus increasing amounts of GFP-CSN6 or Myc-CSN5. The neddylated Cullin-1 proteins were pulled down using Ni-NTA-agarose beads and detected with anti-Nedd8 antibody. The lysates were also immunoprecipitated with CSN6 antibody or CSN5 antibody and were subjected to immunoblotting with anti-Cullin-1. (d) MPN domain of CSN6 had an important role in competing with CSN5 for Cullin-1 binding and affected Cullin-1 neddylation. 293T cells were cotransfected with indicated plasmids. The lysates were immunoprecipitated with anti-Flag or anti-CSN5 ant
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
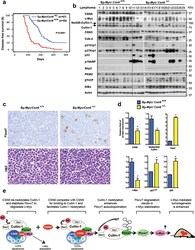
- Figure 6 CSN6 haplo-insufficiency delays the onset/progression of Myc-mediated lymphomagenesis (a) Survival of Eu- Myc mice was prolonged by CSN6 haplo-insufficiency. The genotypes of the transgenic mice are indicated next to the Kaplan-Meier survival curves. The numbers of mice analyzed in each group are denoted. (b) CSN6 haplo-insufficiency caused low levels of Cullin neddylation and low expression of Myc. Lymphoma tissues from Eu- Myc / Csn6 +/+ (1-10) and Eu- Myc / Csn6 +/- (11-25) mice were immunoblotted with the indicated antibodies. (c) Levels of Fbxw7 were elevated in lymphomas from Eu- Myc / Csn6 +/- mice . Lymphomas arising from mice from (b) underwent IHC staining. Representative photographs of Fbxw7 and H&E staining are shown. (d) Levels of neddylated Cullin and Myc were reduced and level of Fbxw7 was increased in lymphomas from transgenic mice. Levels of indicated proteins in lymphomas arising from mice in (b) and (c) were quantitated by integrating optical density from Image J and are demonstrated as bar graphs. Each result shown is representative of 3 independent experiments. Error bars represent 95% CI; Student t-test ,* P< 0.001. (e) The model depicts the pivotal role of the CSN6-Cullin-Fbxw7 axis in Myc-induced lymphomagenesis.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
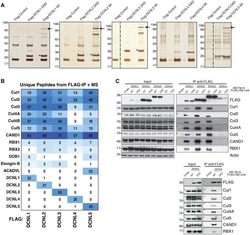
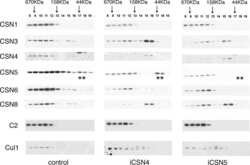
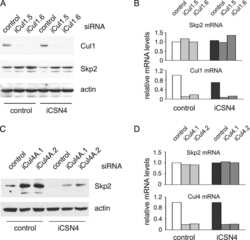
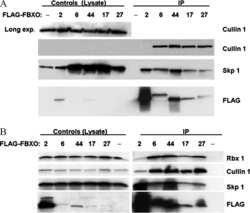
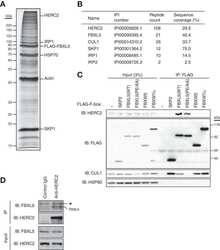
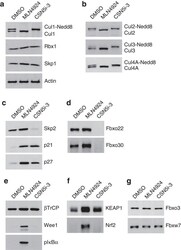
- Figure 2 CSN5 inhibition results in the inactivation of a subset of CRLs and the stabilization of their substrates. Effect on CRL components and substrates after treatment of HCT116 cells with either 1 muM MLN4924 or 1 muM CSN5i-3. ( a ) Immunoblottings for Cul1 and its CRL components Rbx and Skp1; ( b ) for cullins Cul2, Cul3 and Cul4A; ( c ) for F-box protein Skp2 and the SCF Skp2 substrates p21 and p27; ( d ) for F-box proteins Fbxo22 and Fbxo30; ( e ) for F-box protein betaTrCP and the SCF betaTrCP substrates Wee1 and pIkappaBalpha; ( f ) for the adaptor KEAP1 and the Cul3 KEAP1 substrate Nrf2; ( g ) for F-box proteins Fbxw7 and Fbxo3.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
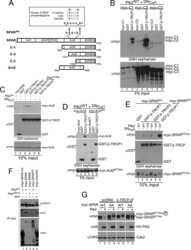
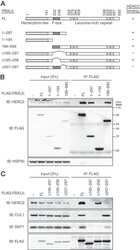
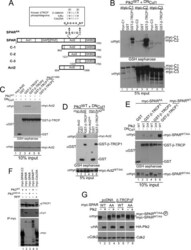
- FIGURE 4. A candidate beta -TRCP phosphodegron in SPAR. A , schematic representation of SPAR domains and SPAR fragments: actin-binding domains ( Act1 and Act2 ), RapGAP, PDZ, and guanylate kinase binding ( GKBD ) domain. The candidate DSGIDT phosphodegron motif (residues 1304-1309) identified in the Act2 domain of SPAR closely resembles the consensus beta-TRCP phosphodegron ( inset ). Boundaries of generated C-terminal fragments of SPAR are depicted; C-1, C-2, and Act2 fragments contain the putative phosphodegron motif, whereas C-3 does not. B and C , SPAR fragments spanning the beta-TRCP phosphodegron bind to beta-TRCP. HEK293T cells were transfected with vectors expressing SPAR fragments C-1, C-2, C-3 ( B ) or Act2 ( C ) (0.6 mug) either alone or with pCMV-GST or pCMV-GST-beta-TRCP (0.6 mug). In addition, pCMV-HA-Plk2 WT/K108M (0.6 mug) and DN Cul1 (2 mug) were co-transfected as indicated. After 24 h, cell extracts were used for GSH-Sepharose pulldown assays, and proteins were immunoblotted with Myc antibodies. Crude lysates were blotted as an input control. D and E , phosphodegron-dependent binding to beta-TRCP. Constructs expressing point mutations (S1305A, T1309A) in full-length SPAR ( E ) and the Act2 fragment ( D ) (myc-SPAR AA or myc-Act2 AA , 0.6 mug) were transfected into HEK293T cells along with pCMV-HA-Plk2 WT/D201A (0.6 mug), pCMV- DN Cul1 (2 mug), and pCMV-GST or pCMV-GST-beta-TRCP (0.6 mug) as indicated. Cell lysates were incubated with GSH-Sepharose and immuno
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
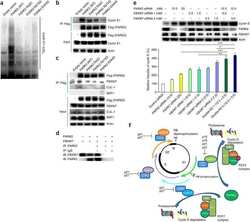
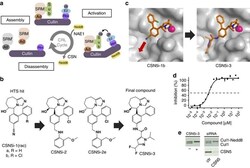
- Figure 1 CSN5i-3 is a potent inhibitor of CSN5-catalysed cullin deneddylation. ( a ) Schematic illustration of the CRL cycle and the role of cullin neddylation and deneddylation. Ad, adaptor protein; SRM, interchangeable substrate recognition module; Sub, substrate. ( b ) Chemical structures of CSN5 inhibitors illustrating the optimization of the high throughput screening (HTS) hit CSN5i-1a to the cell active intermediate CSN5i-2, its R,R -enantiomer CSN5i-2e and to the final compound CSN5i-3. ( c ) Co-crystal structures of CSN5 with CSN5i-1b and CSN5i-3. The structures revealed the direction for compound extension into the substrate binding cleft of CSN5 (red arrow). ( d ) Dose-response curve for inhibition of CSN activity by CSN5i-3 as measured in a biochemical enzyme assay ( n =2, +-s.d.). ( e ) Immunoblotting for Cul1 after treatment of HCT116 cells with 1 muM CSN5i-3 and for Cul1 and CSN5 after treatment with CSN5 siRNA.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
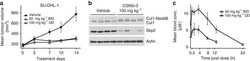
- Figure 4 CSN5i-3 inhibits tumour growth of a human xenograft. ( a ) SU-DHL-1 xenografts were grown in SCID-bg mice and dosed by oral administration with either vehicle control or CSN5i-3 at the indicated doses and schedules. Mean tumour volumes are shown+-s.e.m. ( n =4; P
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
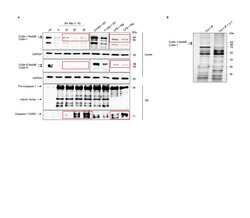
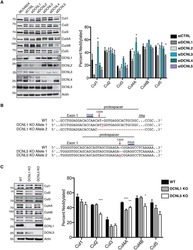
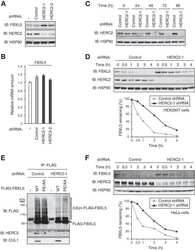
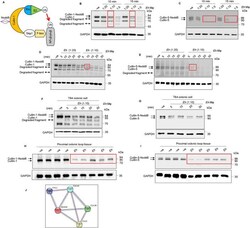
- 10.1371/journal.ppat.1009936.g001 Fig 1 Entamoeba histolytica promotes the degradation of cullin-1/5 from THP-1 derived macrophages, colonic epithelial cells, and proximal colonic loop tissues in a time- and dose-dependent fashion. (A) Schematic representation of Skp-1-Cullin-1-F-box (SCF) protein E3 complex (Skp-1: S-phase kinase-associated protein 1; Rbx: RING [Really Interesting New Gene]-box1; E2: Ubiquitin-loaded E2). (B, C) THP-1 macrophages were incubated with different Eh :macrophage ratios, or (D, E) for increasing times (5 to 30 min) with Eh (20:1 ratio) and Eh (10:1 ratio). (F, G) T84 colonic epithelial cells were grown in 12-well plates and stimulated with Eh (10:1 ratio) for 5 to 30 min. (H, I) Proximal colonic loops were inoculated with Eh and cell lysates were prepared. Post Eh stimulations, cells were washed and cytoplasmic extracts were prepared, and an equal amount of protein was loaded on to the SDS- PAGE gel (7.5%) and immunoblotted with the anti-cullin-1, anti-cullin-5 and anti-GAPDH antibody. Highlighted boxed areas on the figures show point of interest for cullin-1/5 as described in text. (J) Protein-protein interaction using STRING v11 showed direct interaction between cullin-1, cullin-5, cullin-4A, cullin-4B and Nedd8. Data (B, C, D, E, H, and I) are representative of three independent experiments and data (F and G) are representative of two independent experiment and statistical significance was carried out with Students t- test. Bar represent mean +
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- 10.1371/journal.ppat.1009936.g005 Fig 5 Caspase-1 activation during Eh- macrophage contact degrades cullin-1/5. ( A, B) THP-1 derived macrophages were pre-incubated with the pan-caspase inhibitor Z-VAD-fmk (100muM) and caspase-1 specific inhibitor Z-YVAD-fmk (100muM) for 1 h followed by stimulation with Eh (10:1 ratio) for 10 min. Cleavage of the cullin-1/5 were assessed by western blot. (C, D) Wild type (WT) THP-1 and CASP1 CRISPR/Cas9-KO macrophages were stimulated with Eh (10:1 ratio) for 10 min. (E, F) WT, THP-1 def ASC and NLRP3 CRISPR/Cas9-KO THP-1 macrophages were stimulated with Eh (10:1 ratio) for 10 min. After incubation, cells were washed and lysed. Equal amount of protein was loaded on to the SDS-PAGE (7.5%) and immunoblotted with anti-cullin-1 ( Panel: A, C, and E ) , anti-cullin-5 (Panel: B, D and F) and anti-GAPDH antibody. (G) Cullin-1 was immunoprecipitated (Cul-1 IP) using anti cullin-1 antibody and post immunoprecipitation it was incubated with recombinant caspase-1 (C-1) for 4h or overnight (O/N). (H) Immunoprecipitated cullin-1 was incubated with recombinant caspase-1 or with Y-VAD-fmk (50muM or 100muM) or Z-VAD-fmk (100muM). (I) Immunoprecipitated cullin-5 was incubated with recombinant caspase-1 or with Z-VAD-fmk (100muM) or Y-VAD-fmk (100muM). (J) Recombinant cullin-1 (rec Cul-1) was incubated with recombinant caspase-1 overnight at 37deg and was immunoblotted with anti-cullin-1 antibody. Note, recombinant cullin-1 (rec Cul-1) has a molecular weight of
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
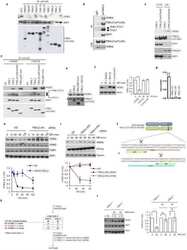
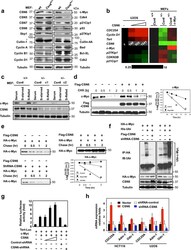
- Figure 1 Loss of CSN6 leads to destabilization of Myc and decreases Myc transcriptional activity (a) Immunoblot analysis of Csn6 +/- MEFs. Primary MEF cells were prepared from 13.5-day embryos derived from wild-type (wt) Csn6 +/+ and Csn6 +/- mice. After 4 days of culture, cell lysates were analyzed by indicated antibodies. (b) Quantitative RT-PCR analysis of mRNA levels for indicated c-Myc target genes in CSN6-transfected U2OS cells or Csn6 +/- MEFs. The quantitated mRNA expression level was normalized to GAPDH mRNA. Two MEFs were examined for each genotype. Expression level of transfected CSN6 is indicated as numbers on the heat map, and the heat map depicts the natural logarithm of fold-change in mRNA expression. (c) CSN6 knockdown abrogated serum-induced elevation of Myc. Csn6 +/+ MEFs and Csn6 +/- clone 1 (C1) and clone 2 (C2) MEFs were serum-starved for 24 hours and then were refed with serum for 24 hours. Lysates were analyzed by indicated antibodies. (d) CSN6 decreased Myc turnover. 293T cells were transfected with Flag-CSN6 and were then treated with cycloheximide (CHX; 100 ug/ml) for the indicated times. The immunoblot of Myc signal at each time point was measured using a densitometer and the integrated optical density of Myc was measured. The turnover of Myc is indicated graphically. (e) CSN6 expression affected Myc turnover. 35 S-pulse-labeled HA-Myc protein was immunoprecipitated from indicated transfected 293T cell lysates. The mixture was separated by sodium do
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Fig. 4 Endogenic Cul1 deneddylation is enhanced during simultaneous neddylation. a 20 mug HeLa cells extract were incubated for 30 min at 30degC ( lanes 2 - 7 ) or with the addition of ATP-trap mix prior to the incubation ( lanes 8 - 13 ). Simultaneous ATP-depletion was performed in lanes 3 - 7 after 30 min incubation, on a time plot of 1.5-7.5 min at 30degC. Simultaneous neddylation was performed in lanes 9 - 13 after 30 min, by the addition of AMP-PNP, on a time plot of 1.5-7.5 min at 30degC. b 10 nM His-6-Cul1/ROC1 was neddylated at 30degC on a time plot of 2.5-30 min, without or with the addition of ATP-trap mix ( lanes 2 - 6 and lanes 7 - 11 , respectively). In lane 12 , the neddylation mix was added to the reaction after pre-incubation with ATP-trap mix. c 10 nM His-6-Cul1/ROC1 was neddylated at 30degC using AMP-PNP on a time plot of 2.5-30 min, either directly ( lanes 2 - 6 ) or after pre-incubation with ATP-trap mix ( lanes 7 - 11 ).
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot ELISA
ELISA Flow cytometry
Flow cytometry