Antibody data
- Antibody Data
- Antigen structure
- References [23]
- Comments [0]
- Validations
- Western blot [3]
- Immunoprecipitation [1]
- Immunohistochemistry [5]
- Flow cytometry [1]
- Other assay [30]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 51-1800 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- Cullin 2 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Synthetic peptide
- Reactivity
- Human, Mouse, Rat
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μg
- Concentration
- 0.25 mg/mL
- Storage
- -20°C
Submitted references Cullin neddylation inhibitor attenuates hyperglycemia by enhancing hepatic insulin signaling through insulin receptor substrate stabilization.
Quantitative analyses for effects of neddylation on CRL2(VHL) substrate ubiquitination and degradation.
Desmosomal COP9 regulates proteome degradation in arrhythmogenic right ventricular dysplasia/cardiomyopathy.
Inducible Degradation of Target Proteins through a Tractable Affinity-Directed Protein Missile System.
Small molecule perturbation of the CAND1-Cullin1-ubiquitin cycle stabilizes p53 and triggers Epstein-Barr virus reactivation.
An affinity-directed protein missile system for targeted proteolysis.
Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle.
Oxygen-dependent Regulation of Erythropoietin Receptor Turnover and Signaling.
Ubiquitin-conjugating enzyme Cdc34 and ubiquitin ligase Skp1-cullin-F-box ligase (SCF) interact through multiple conformations.
Ubiquitin-conjugating enzyme Cdc34 and ubiquitin ligase Skp1-cullin-F-box ligase (SCF) interact through multiple conformations.
CCDC22 deficiency in humans blunts activation of proinflammatory NF-κB signaling.
DCNL1 functions as a substrate sensor and activator of cullin 2-RING ligase.
The P-body component USP52/PAN2 is a novel regulator of HIF1A mRNA stability.
UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1α accumulation.
NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway.
COMMD1 (copper metabolism MURR1 domain-containing protein 1) regulates Cullin RING ligases by preventing CAND1 (Cullin-associated Nedd8-dissociated protein 1) binding.
Neddylation-induced conformational control regulates cullin RING ligase activity in vivo.
Characterization of the role of COP9 signalosome in regulating cullin E3 ubiquitin ligase activity.
GCN5 is a required cofactor for a ubiquitin ligase that targets NF-kappaB/RelA.
Substrate-mediated regulation of cullin neddylation.
Renal cell carcinoma risk in type 2 von Hippel-Lindau disease correlates with defects in pVHL stability and HIF-1alpha interactions.
Tumor suppression by the von Hippel-Lindau protein requires phosphorylation of the acidic domain.
SOCS-1 localizes to the microtubule organizing complex-associated 20S proteasome.
Chen C, Gu L, Matye DJ, Clayton YD, Hasan MN, Wang Y, Friedman JE, Li T
Proceedings of the National Academy of Sciences of the United States of America 2022 Feb 8;119(6)
Proceedings of the National Academy of Sciences of the United States of America 2022 Feb 8;119(6)
Quantitative analyses for effects of neddylation on CRL2(VHL) substrate ubiquitination and degradation.
Wang K, Reichermeier KM, Liu X
Protein science : a publication of the Protein Society 2021 Nov;30(11):2338-2345
Protein science : a publication of the Protein Society 2021 Nov;30(11):2338-2345
Desmosomal COP9 regulates proteome degradation in arrhythmogenic right ventricular dysplasia/cardiomyopathy.
Liang Y, Lyon RC, Pellman J, Bradford WH, Lange S, Bogomolovas J, Dalton ND, Gu Y, Bobar M, Lee MH, Iwakuma T, Nigam V, Asimaki A, Scheinman M, Peterson KL, Sheikh F
The Journal of clinical investigation 2021 Jun 1;131(11)
The Journal of clinical investigation 2021 Jun 1;131(11)
Inducible Degradation of Target Proteins through a Tractable Affinity-Directed Protein Missile System.
Simpson LM, Macartney TJ, Nardin A, Fulcher LJ, Röth S, Testa A, Maniaci C, Ciulli A, Ganley IG, Sapkota GP
Cell chemical biology 2020 Sep 17;27(9):1164-1180.e5
Cell chemical biology 2020 Sep 17;27(9):1164-1180.e5
Small molecule perturbation of the CAND1-Cullin1-ubiquitin cycle stabilizes p53 and triggers Epstein-Barr virus reactivation.
Tikhmyanova N, Tutton S, Martin KA, Lu F, Kossenkov AV, Paparoidamis N, Kenney S, Salvino JM, Lieberman PM
PLoS pathogens 2017 Jul;13(7):e1006517
PLoS pathogens 2017 Jul;13(7):e1006517
An affinity-directed protein missile system for targeted proteolysis.
Fulcher LJ, Macartney T, Bozatzi P, Hornberger A, Rojas-Fernandez A, Sapkota GP
Open biology 2016 Oct;6(10)
Open biology 2016 Oct;6(10)
Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle.
Mosadeghi R, Reichermeier KM, Winkler M, Schreiber A, Reitsma JM, Zhang Y, Stengel F, Cao J, Kim M, Sweredoski MJ, Hess S, Leitner A, Aebersold R, Peter M, Deshaies RJ, Enchev RI
eLife 2016 Mar 31;5
eLife 2016 Mar 31;5
Oxygen-dependent Regulation of Erythropoietin Receptor Turnover and Signaling.
Heir P, Srikumar T, Bikopoulos G, Bunda S, Poon BP, Lee JE, Raught B, Ohh M
The Journal of biological chemistry 2016 Apr 1;291(14):7357-72
The Journal of biological chemistry 2016 Apr 1;291(14):7357-72
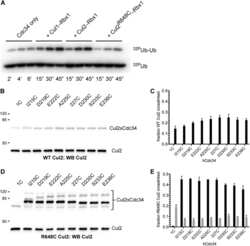
Ubiquitin-conjugating enzyme Cdc34 and ubiquitin ligase Skp1-cullin-F-box ligase (SCF) interact through multiple conformations.
Sandoval D, Hill S, Ziemba A, Lewis S, Kuhlman B, Kleiger G
The Journal of biological chemistry 2015 Jan 9;290(2):1106-18
The Journal of biological chemistry 2015 Jan 9;290(2):1106-18
Ubiquitin-conjugating enzyme Cdc34 and ubiquitin ligase Skp1-cullin-F-box ligase (SCF) interact through multiple conformations.
Sandoval D, Hill S, Ziemba A, Lewis S, Kuhlman B, Kleiger G
The Journal of biological chemistry 2015 Jan 9;290(2):1106-18
The Journal of biological chemistry 2015 Jan 9;290(2):1106-18
CCDC22 deficiency in humans blunts activation of proinflammatory NF-κB signaling.
Starokadomskyy P, Gluck N, Li H, Chen B, Wallis M, Maine GN, Mao X, Zaidi IW, Hein MY, McDonald FJ, Lenzner S, Zecha A, Ropers HH, Kuss AW, McGaughran J, Gecz J, Burstein E
The Journal of clinical investigation 2013 May;123(5):2244-56
The Journal of clinical investigation 2013 May;123(5):2244-56
DCNL1 functions as a substrate sensor and activator of cullin 2-RING ligase.
Heir P, Sufan RI, Greer SN, Poon BP, Lee JE, Ohh M
Molecular and cellular biology 2013 Apr;33(8):1621-31
Molecular and cellular biology 2013 Apr;33(8):1621-31
The P-body component USP52/PAN2 is a novel regulator of HIF1A mRNA stability.
Bett JS, Ibrahim AF, Garg AK, Kelly V, Pedrioli P, Rocha S, Hay RT
The Biochemical journal 2013 Apr 15;451(2):185-94
The Biochemical journal 2013 Apr 15;451(2):185-94
UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1α accumulation.
Bandau S, Knebel A, Gage ZO, Wood NT, Alexandru G
BMC biology 2012 Apr 26;10:36
BMC biology 2012 Apr 26;10:36
NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway.
den Besten W, Verma R, Kleiger G, Oania RS, Deshaies RJ
Nature structural & molecular biology 2012 Apr 1;19(5):511-6, S1
Nature structural & molecular biology 2012 Apr 1;19(5):511-6, S1
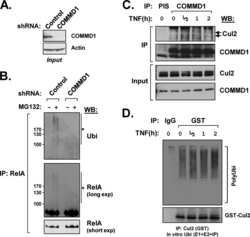
COMMD1 (copper metabolism MURR1 domain-containing protein 1) regulates Cullin RING ligases by preventing CAND1 (Cullin-associated Nedd8-dissociated protein 1) binding.
Mao X, Gluck N, Chen B, Starokadomskyy P, Li H, Maine GN, Burstein E
The Journal of biological chemistry 2011 Sep 16;286(37):32355-65
The Journal of biological chemistry 2011 Sep 16;286(37):32355-65
Neddylation-induced conformational control regulates cullin RING ligase activity in vivo.
Boh BK, Smith PG, Hagen T
Journal of molecular biology 2011 Jun 3;409(2):136-45
Journal of molecular biology 2011 Jun 3;409(2):136-45
Characterization of the role of COP9 signalosome in regulating cullin E3 ubiquitin ligase activity.
Choo YY, Boh BK, Lou JJ, Eng J, Leck YC, Anders B, Smith PG, Hagen T
Molecular biology of the cell 2011 Dec;22(24):4706-15
Molecular biology of the cell 2011 Dec;22(24):4706-15
GCN5 is a required cofactor for a ubiquitin ligase that targets NF-kappaB/RelA.
Mao X, Gluck N, Li D, Maine GN, Li H, Zaidi IW, Repaka A, Mayo MW, Burstein E
Genes & development 2009 Apr 1;23(7):849-61
Genes & development 2009 Apr 1;23(7):849-61
Substrate-mediated regulation of cullin neddylation.
Chew EH, Hagen T
The Journal of biological chemistry 2007 Jun 8;282(23):17032-40
The Journal of biological chemistry 2007 Jun 8;282(23):17032-40
Renal cell carcinoma risk in type 2 von Hippel-Lindau disease correlates with defects in pVHL stability and HIF-1alpha interactions.
Knauth K, Bex C, Jemth P, Buchberger A
Oncogene 2006 Jan 19;25(3):370-7
Oncogene 2006 Jan 19;25(3):370-7
Tumor suppression by the von Hippel-Lindau protein requires phosphorylation of the acidic domain.
Lolkema MP, Gervais ML, Snijckers CM, Hill RP, Giles RH, Voest EE, Ohh M
The Journal of biological chemistry 2005 Jun 10;280(23):22205-11
The Journal of biological chemistry 2005 Jun 10;280(23):22205-11
SOCS-1 localizes to the microtubule organizing complex-associated 20S proteasome.
Vuong BQ, Arenzana TL, Showalter BM, Losman J, Chen XP, Mostecki J, Banks AS, Limnander A, Fernandez N, Rothman PB
Molecular and cellular biology 2004 Oct;24(20):9092-101
Molecular and cellular biology 2004 Oct;24(20):9092-101
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
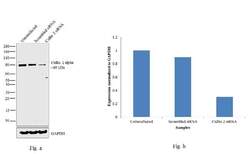
- Experimental details
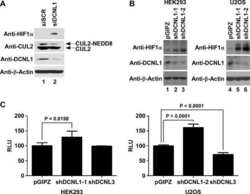
- Knockdown of Cullin 2 was achieved by transfecting NTERA-2 cells with Cullin 2 specific validated siRNAs (Silencer® select Product # s16053, s16051). Western blot analysis (Fig. a) was performed using whole cell extracts from the Cullin 2 knockdown cells (lane 3), non-specific scrambled siRNA transfected cells (lane 2) and untransfected cells (lane 1). The blots were probed with Cullin 2 Polyclonal Antibody (Product # 51-1800, 1 µg/mL) and Goat anti-Rabbit IgG (Heavy Chain) Superclonal™ Secondary Antibody, HRP conjugate (Product # A27036, 0.25 µg/mL, 1:4000 dilution). Densitometric analysis of this western blot is shown in histogram (Fig. b). Decrease in signal upon siRNA mediated knock down confirms that antibody is specific to Cullin 2.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
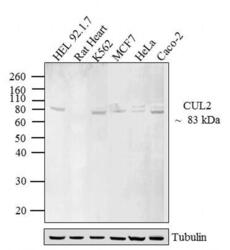
- Experimental details
- Western blot analysis of CUL-2 was performed by loading 20 µg of HEL 92.1.7 (lane1), Rat Heart (lane2), K562 (lane3), MCF7 (lane4), HeLa (lane5) and Caco-2 (lane6) lysate using Novex® NuPAGE® 12 % Bis-Tris gel (Product # NP0342BOX), XCell SureLock™ Electrophoresis System (Product # EI0002), Novex® Sharp Pre-Stained Protein Standard (LC5800), and iBlot® 2 Dry Blotting System (IB21001). Proteins were transferred to a nitrocellulose membrane and blocked with 5 % skim milk for 1 hour at room temperature. CUL-2 was detected at ~ 83 kDa using CUL-2 Rabbit polyclonal Antibody (Product # 51-1800) at 2-3 µg/mL in 5 % skim milk at 4ºC overnight on a rocking platform. Goat Anti-Rabbit IgG - HRP Secondary Antibody (G21234) at 1:5000 dilution was used and chemiluminescent detection was performed using Pierce™ ECL Western Blotting Substrate (Product # 32106).
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
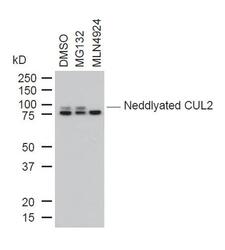
- Experimental details
- Western blot analysis of Cullin-2 was performed by loading 10 µg of whole cell lysates from DMSO, MG132 or MLN4924 treated U2OS cells onto a SDS PAGE gel. Proteins were transferred, blocked and endogenous Cullin-2, as well as neddylated Cullin-2 were detected using a CUL-2 polyclonal antibody (Product # 51-1800) at a dilution of 1:1000.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
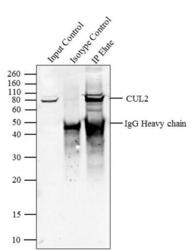
- Main image
- Experimental details
- Immunoprecipitation of CUL2 was performed with 5 µg of the CUL2 Rabbit Polyclonal Antibody (Product # 51-1800) on cell extract from HEL 92.1.7 using the Dynabeads® Protein A Immunoprecipitation Kit (10006D). Normal Rabbit IgG was used as a negative IP control. Subsequently, western blot analysis was performed using Novex® NuPAGE® 4-12 % Bis-Tris gel (Product # NP0321BOX), XCell SureLock™ Electrophoresis System (Product # EI0002), Novex® Sharp Pre-Stained Protein Standard (LC5800). Proteins were transferred using iBlot® 2 Dry Blotting System (IB21001) to a nitrocellulose membrane and blocked with 5% skim milk for 1 hour at room temperature on a rocking platform. CUL2 was detected at ~ 80 kDa using CUL2 Rabbit Polyclonal Antibody (Product # 51-1800) at 2-3 µg/mL in 5 % skim milk at 37ºC for 2 hours. Goat Anti-Rabbit IgG - HRP Secondary Antibody (G21234) at 1:5000 dilution was used and chemiluminescent detection was performed using Pierce™ ECL Western Blotting Substrate (Product # 32106). Lane 1 represents 10 % of the total cell extract (input), Lane 2 is the IP performed with Rabbit IgG and Lane 3 represents IP performed with CUL2 Rabbit Polyclonal Antibody (Product # 51-1800).
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
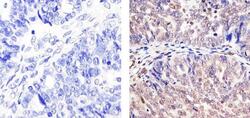
- Experimental details
- Immunohistochemistry analysis of CUL-2 showing staining in the cytoplasm and nucleus of paraffin-embedded human Ovarian carcinoma tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a CUL-2 Rabbit Polyclonal antibody (Product # 51-1800) diluted in 3% BSA-PBS at a dilution of 1:20 overnight at 4ºC in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
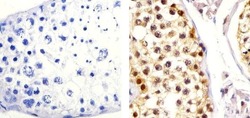
- Experimental details
- Immunohistochemistry analysis of CUL-2 showing staining in the cytoplasm and nucleus of paraffin-embedded human testis tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a CUL-2 Rabbit Polyclonal antibody (Product # 51-1800) diluted in 3% BSA-PBS at a dilution of 1:20 overnight at 4ºC in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
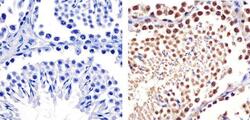
- Experimental details
- Immunohistochemistry analysis of CUL-2 showing staining in the cytoplasm and nucleus of paraffin-embedded Mouse testis tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a CUL-2 Rabbit Polyclonal antibody (Product # 51-1800) diluted in 3% BSA-PBS at a dilution of 1:20 overnight at 4ºC in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
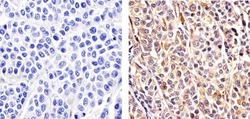
- Experimental details
- Immunohistochemistry analysis of CUL-2 showing staining in the cytoplasm and nucleus of paraffin-embedded human melanoma tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a CUL-2 Rabbit Polyclonal antibody (Product # 51-1800) diluted in 3% BSA-PBS at a dilution of 1:20 overnight at 4ºC in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
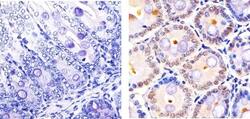
- Experimental details
- Immunohistochemistry analysis of CUL-2 showing staining in the cytoplasm and nucleus of paraffin-embedded Mouse Colon tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a CUL-2 Rabbit Polyclonal antibody (Product # 51-1800) diluted in 3% BSA-PBS at a dilution of 1:20 overnight at 4ºC in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
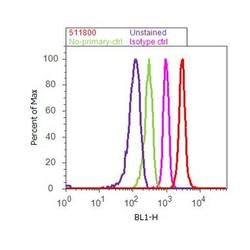
- Experimental details
- Flow cytometry analysis of CUL-2 was done on K-562 cells. Cells were fixed with 70% ethanol for 10 minutes, permeabilized with 0.25% Triton™ X-100 for 20 minutes, and blocked with 5% BSA for 30 minutes at room temperature. Cells were labeled with CUL-2 Rabbit Polyclonal Antibody (511800, red histogram) or with rabbit isotype control (pink histogram) at 3-5 ug/million cells in 2.5% BSA. After incubation at room temperature for 2 hours, the cells were labeled with Alexa Fluor® 488 Goat Anti-Rabbit Secondary Antibody (A11008) at a dilution of 1:400 for 30 minutes at room temperature. The representative 10,000 cells were acquired and analyzed for each sample using an Attune® Acoustic Focusing Cytometer. The purple histogram represents unstained control cells and the green histogram represents no-primary-antibody control.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
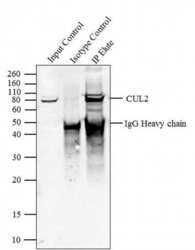
- Experimental details
- Immunoprecipitation of CUL2 was performed with 5 æg of the CUL2 Rabbit Polyclonal Antibody (Product # 51-1800) on cell extract from HEL 92.1.7 using the Dynabeads® Protein A Immunoprecipitation Kit (10006D). Normal Rabbit IgG was used as a negative IP control. Subsequently, western blot analysis was performed using Novex® NuPAGE® 4-12 % Bis-Tris gel (Product # NP0321BOX), XCell SureLock™ Electrophoresis System (Product # EI0002), Novex® Sharp Pre-Stained Protein Standard (LC5800). Proteins were transferred using iBlot® 2 Dry Blotting System (IB21001) to a nitrocellulose membrane and blocked with 5% skim milk for 1 hour at room temperature on a rocking platform. CUL2 was detected at ~ 80 kDa using CUL2 Rabbit Polyclonal Antibody (Product # 51-1800) at 2-3 æg/mL in 5 % skim milk at 37ºC for 2 hours. Goat Anti-Rabbit IgG - HRP Secondary Antibody (G21234) at 1:5000 dilution was used and chemiluminescent detection was performed using Pierce™ ECL Western Blotting Substrate (Product # 32106). Lane 1 represents 10 % of the total cell extract (input), Lane 2 is the IP performed with Rabbit IgG and Lane 3 represents IP performed with CUL2 Rabbit Polyclonal Antibody (Product # 51-1800).
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
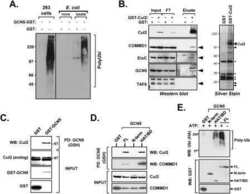
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
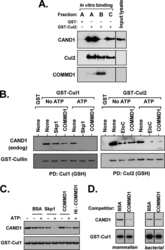
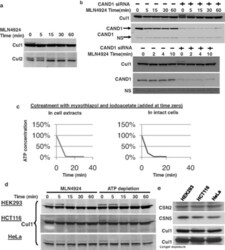
- FIGURE 2: In vivo cullin deneddylation rates in the presence and absence of ongoing substrate ubiquitination. (A) HEK293 cells were grown in 12-well plates and 3 muM MLN4924 was added at time zero. Cells were lysed at the indicated time points and cell lysates were analyzed by Western blotting with Cul1 and Cul2 antibodies. (B) Cells were transfected with negative control or CAND1 siRNA for 3 d. The Cul1 deneddylation rate was determined as in (A). NS, nonspecific band that served as a loading control. (C) Cells were cotreated with 1 muM myxothiazol and 2.5 mM iodoacetate to rapidly deplete cellular ATP concentrations. ATP concentrations were measured as described in Materials and Methods . (D) To measure the Cul1 deneddylation rate in the presence and absence of ongoing substrate ubiquitination, HEK293, HCT116, and HeLa cells were treated with either MLN-4924 (3 muM) or myxothiazol (1 muM) and iodoacetate (2.5 mM) at time zero. Cells were lysed at the indicated time points and cell lysates were analyzed by Western blotting with Cul1 antibody. The Western blots shown are representative of at least three independent experiments in each cell line and did not show any consistent difference in the rate at which neddylation occurs. (E) The relative expression levels of CSN and Cul1 in HEK293, HCT116, and HeLa cells was compared by Western blotting of equal protein amounts of cell lysate with the indicated antibodies.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
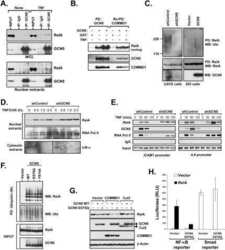
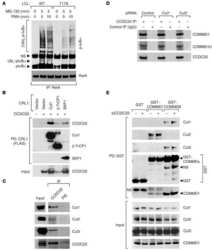
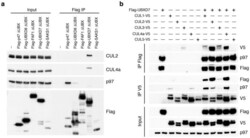
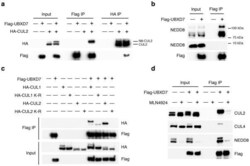
- Figure 1 UBXD7 associates with all cullins except CUL5 (a) Flag-tagged UBA-UBX proteins (p47, UBXD8, FAF1, UBXD7 and SAKS1) lacking the UBX domain (DeltaUBX) expressed in 293T cells were immunoprecipitated (IP) with anti-Flag antibodies and probed to detect endogenous binding partners CUL2, CUL4, and p97 as indicated. -, no transfection. (b) Same as in a, except cells were transfected with (+) or without (-) vectors encoding Flag-UBXD7 and V5 epitope-tagged CUL 1 through 5. Immunoprecipitations were carried out with antibodies against the Flag or V5 epitope.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- Figure 2 The UBXD7-CRL interaction is neddylation dependent (a-b) 293T cells were transfected with (+) or without (-) the indicated expression constructs and immunoprecipitated (IP) with anti-Flag or anti-HA antibodies prior to being probed with the antibodies indicated. N8-CUL2, NEDD8-conjugated CUL2. (c) Same as in a. HA-CUL1 (K/R) contains the K720R substitution and HA- CUL2 (K/R) contains the K689R substitution. (d) Same as above, except 1 hour prior to cell lysis, cells were treated with the NEDD8 conjugation inhibitor, MLN4924.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
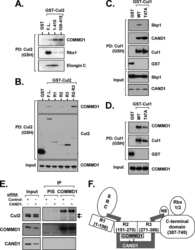
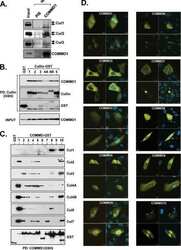
- Figure 3 UBXD7 directly interacts with neddylated CRLs via its UIM (a) Wild type UBXD7 protein, indicating its domains. UBA, ubiquitin-associated domain, UAS, UAS/Thioredoxin-like fold domain, UIM, ubiquitin-interacting motif, UBX, ubiquitin regulatory X domain. (b) Recombinant wild-type Flag-UBXD7 or a deletion mutant was incubated with K48-linked polyubiquitin chains prior to immunoprecipitation and western blotting with indicated antibodies. N x Ub refers to ubiquitin chains of increasing length. (c) A mix (input) of unneddylated and neddylated recombinant full-length CUL2-RBX1 or CUL4a-RBX1 complex was incubated with recombinant WT Flag-tagged UBXD7 or deletion mutants. Following IP with anti-Flag antibodies, recovered proteins were detected by western blotting with indicated antibodies. N x N8-CUL, cullin modified with 1 or 2 molecules of NEDD8. (d) 293T cells were transfected with full length Flag-tagged UBXD7 or the indicated UBXD7 deletion mutants and treated with MG132. Lysates were immunoprecipitated with anti-Flag antibodies, and co-precipitated endogenous proteins were detected by western blotting with the indicated antibodies. (e) Binding assays were performed as in (c) using a mix of unneddylated and neddylated recombinant CUL2 CTD-RBX1 or CUL2 CTD-RBX2 complex and recombinant WT Flag-tagged UBXD7 or deletion mutants.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
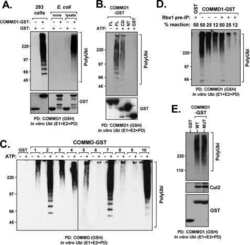
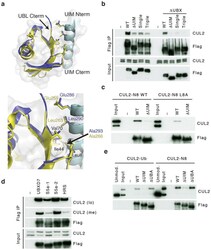
- Figure 4 The UIM of UBXD7 binds conjugated NEDD8 on CUL2 (a) Modeling of the UBXD7 UIM - NEDD8 interaction using the HRS UIM -ubiquitin crystal structure as a template (PDB code 2D3G, yellow) 17 . The UBXD7 UIM in association with NEDD8 is shown in blue. This figure was made in PYMOL. (b) Cells transfected with Flag-UBXD7 (wild type or DeltaUBX) with a wild type, deleted (DeltaUIM), or substitution mutant UIM were immunoprecipitated with anti-Flag beads and probed with the indicated antibodies. (c) CUL2 was neddylated with wild type (WT) NEDD8 or L8A mutant NEDD8 and binding assays with Flag-UBXD7 were performed as described in Fig. 3c . (d) Same as in (c), except transfections were carried out with wild type Flag-UBXD7 or Flag-UBXD7 in which the UIM was replaced with the UIM of HRS or the first (S5a-1) or second (S5a-2) UIM of S5a. Immunoprecipitations were carried out in either a low (CUL2 (lo)) or medium (CUL2 (me)) stringency binding buffer. (e) Immunoprecipitation of recombinant wild type Flag-tagged UBXD7 or the indicated UBXD7 deletion mutants from a mix (input) containing recombinant full-length CUL2-RBX1 either unmodified or modified with monoubiquitin or NEDD8.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
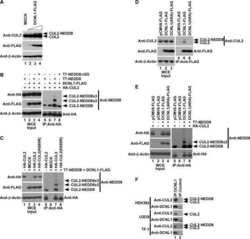
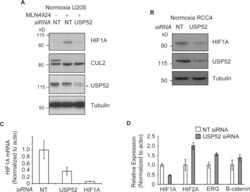
- Figure 3 USP52 regulates the hypoxia pathway in a VHL-independent manner by controlling HIF1A mRNA levels ( A ) Normoxic U2OS cells were treated with MLN4924 to block CUL2 NEDD8ylation andinactivate the VHL complex. USP52 depletion was sufficient to decrease HIF1A protein levels,demonstrating independence of the VHL complex. Tubulin was used as a loading control.( B ) USP52 depletion in VHL-deficient RCC4 renal cancer cells reduced HIF1A proteinlevels under normoxia. Tubulin was used as a loading control. ( C ) U2OS cells weretreated with siRNA against USP52 and HIF1A , and the levels of HIF1A mRNA were assessed by real-time RT-PCR analysis. HIF1A siRNA reduced HIF1A mRNA levels to below 10%, whereas USP52depletion reduced HIF1A mRNA to approximately 40%. Levels were normalized tobeta-actin. ( D ) USP52 was depleted in HeLa cells which were subject to real-timeRT-PCR analysis. HIF1A mRNA levels were reduced by approximately 50% in HeLacells by USP52 knockdown. HIF2A levels were increased 2-fold upon USP52 knockdown, whereas ERG and CTNNB1 mRNAlevels were both increased 1.5-fold upon USP52 depletion. Levels were normalized to beta-actin.Molecular masses are indicated in kDa in the blots, and results in histograms aremeans+-S.E.M.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
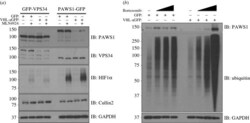
- Experimental details
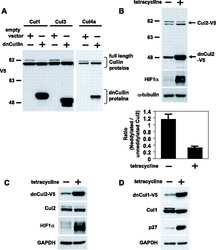
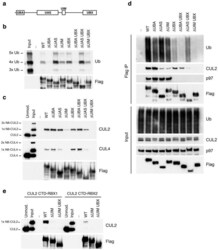
- Figure 2. Inhibition of Cullin Neddylation and the proteasome rescues protein degradation through AdPROM: ( a ) retroviral infections of HEK293 and U2OS cells, harbouring GFP-VPS34 and PAWS1-GFP knockins, respectively, were performed to allow the expression of either the control (GFP) or VHL-aGFP polypeptide. Infected cells were then treated with either DMSO or the pan-Cullin Neddylation inhibitor MLN4924 (1 uM) for 6 h, as indicated. Extracts (20 ug protein) were resolved by SDS-PAGE, transferred to PVDF membranes and subjected to western blotting for PAWS1, VPS34, HIF1alpha and Cullin2 expression as shown. GAPDH was included as a loading control. ( b ) Control (GFP) or VHL-aGFP PAWS1-GFP cells described in ( a ) were treated with 10 uM Bortezomib for 0, 4, 10 or 24 h prior to lysis. Extracts (20 ug protein) were resolved by SDS-PAGE, transferred to PVDF membranes and subjected to western blotting using antibodies against PAWS1, total ubiquitin and GAPDH control as indicated.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
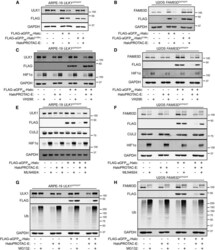
- Experimental details
- Figure 2 Characterization of HaloPROTAC-E L-AdPROM-Mediated GFP-ULK1 and FAM83D-GFP Degradation (A and B) ARPE-19 ULK1 GFP/GFP (A) or U2OS FAM83D GFP/GFP (B) FLAG-empty, FLAG-aGFP 6M -Halo and FLAG-aGFP 6M -Halo D106A binding mutant-expressing cells were treated with 250 nM (A) or 1 muM (B) HaloPROTAC-E for 24 h. (C and D) ARPE-19 ULK1 GFP/GFP (C) or U2OS FAM83D GFP/GFP (D) FLAG-empty and FLAG-aGFP 6M -Halo-expressing cells were treated with 250 nM (C) or 1 muM (D) HaloPROTAC-E and 50 muM VHL inhibitor VH298 for 24 h. (E and F) ARPE-19 ULK1 GFP/GFP (E) or U2OS FAM83D GFP/GFP (F) FLAG-empty and FLAG-aGFP 6M -Halo-expressing cells were treated with 250 nM (E) or 1 muM (F) HaloPROTAC-E and 1 muM pan-Cullin NEDDylation inhibitor MLN4924 for 24 h. (G and H) ARPE-19 ULK1 GFP/GFP (G) or U2OS FAM83D GFP/GFP (H) FLAG-empty and FLAG-aGFP 6M -Halo-expressing cells were treated with 250 nM (G) or 1 muM (H) HaloPROTAC-E and 20 muM proteasome inhibitor MG132 for 24 h. For (A)-(H), extracts were resolved by SDS-PAGE and transferred on to PVDF membranes, which were subjected to immunoblotting with indicated antibodies.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
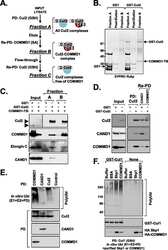
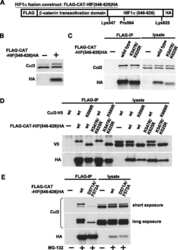
- Fig 6 ZTA transcription activation domain mediates interactions with CAND1 and Cullins. (A) Schematic of ZTA transcription activation (TA) indicating position of sumoylation site at K12 and Cul5 box 52-LPEP-54. (B) Luciferase assay for BHLF1-Luc cotransfected with expression vector for ZTA (WT), K12A, or EP53/54AA (MUT) in 293T cells treated with DMSO (black bars) or C60 (grey bars) for 48 hrs. (C) Western blot of 293T cells transfected with ZTA (WT), K12A, or EP53/54AA treated with DMSO or C60 for 48 hrs, as described in panel B. (D-E) 293T cells transfected with FL-CTRL, FL-WT-ZTA, or FL-MUT-ZTA were treated with DMSO or C60 for 48 hrs, and then subject to IP with FLAG and probed with antibodies for Cul 1, Cul 2, CAND1, FLAG, or Actin (D) . 10% of input was analyzed by Western for Cul 1, Cul 2, CAND1, FLAG, and Actin (E) . (F-G) 293T cells were transfected as in D-E, and subject to IP with either Cul 1, Cul 2, or FLAG, and then analyzed by Western blot with antibody to FLAG (F) . 10% of input was analyzed by Western for Cul 1, Cul 2, FLAG, and Actin (G) .
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
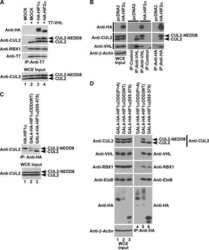
- FIGURE 1 Eliminating cullin neddylation failed to inhibit the degradation of HIF1alpha in HEK293 cells. (a) HEK293 cells were pretreated with DFX for 5 h and pevonedistat or equivalent volume of DMSO for 1 h. After washout of inhibitors, cells were treated by CHX with or without pevonedistat for indicated time and were collected for WB analyses. (b) same as in (a) except that HEK293 cells containing tetracycline induced FLAG HIF1alpha were used
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
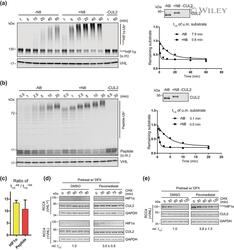
- Experimental details
- 2 FIGURE Neddylation is required for efficient CRL2 VHL -dependent degradation of HIF1alpha in vitro and in RCC4 (+VHL) cells. (a,b) Full-length FLAG HIF1alpha purified from RCC4 (VHL ) cells stably expressing FLAG HIF1alpha (a) or HIF1alpha degron peptide (b) were ubiquitinated in vitro for indicated time with or without CUL2 neddylation. Samples without CUL2 served as negative controls. Intensities of unmodified (u.m.) FLAG HIF1alpha or degron peptide bands were normalized to intensities of VHL bands for regression analyses. WB of CUL2 from each group and regression curves of remaining unmodified substrates were shown on the right. (c) Average relative changes of substrate t 1/2 by neddylation from experiments in (a,b). Error bars: range of values, n = 2. (D) RCC4 (VHL ) and RCC4 (+VHL) cells were pretreated with DFX for 3 h and pevonedistat or equivalent volume of DMSO for 1 h. After washout of inhibitors, cells were treated by CHX with or without pevonedistat for indicated time and were collected for WB analyses. (e) Same assays as performed in (d) with FLAG HIF1alpha stably expressed in RCC4 (+VHL) cells. In (d,e), relative t 1/2 was shown as average +- range of values ( n = 2)
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot ELISA
ELISA