Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Immunohistochemistry [3]
- Other assay [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-59337 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- GNAO1 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant protein fragment
- Description
- Immunogen sequence: IQSLAAIVRA MDTLGIEYGD KERKADAKMV CDVVSRMEDT EPFSAELLSA MMRLWGDSGI QECF Highest antigen sequence identity to the following orthologs: Mouse - 95%, Rat - 97%.
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 0.1 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references Notch signaling determines cell-fate specification of the two main types of vomeronasal neurons of rodents.
Katreddi RR, Taroc EZM, Hicks SM, Lin JM, Liu S, Xiang M, Forni PE
Development (Cambridge, England) 2022 Jul 1;149(13)
Development (Cambridge, England) 2022 Jul 1;149(13)
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
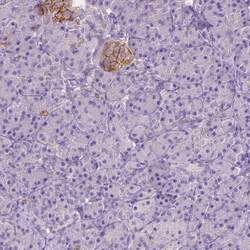
- Experimental details
- Immunohistochemical staining of GNAO1 in human pancreas using GNAO1 Polyclonal Antibody (Product # PA5-59337) shows low expression as expected.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
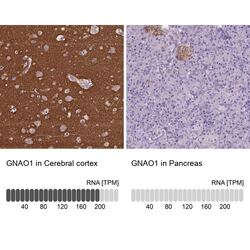
- Experimental details
- Immunohistochemical staining of GNAO1 in human cerebral cortex and pancreas tissues using GNAO1 Polyclonal Antibody (Product # PA5-59337). Corresponding GNAO1 RNA-seq data are presented for the same tissues.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Immunohistochemical staining of GNAO1 in human cerebral cortex using GNAO1 Polyclonal Antibody (Product # PA5-59337) shows high expression.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
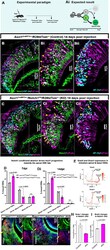
- Experimental details
- Fig. 5. Notch1 loss of function biases VSN differentiation to the apical fate. (A) Ascl1Cre ERT2 /Notch1 fl/fl /R26tdTom and Ascl1Cre ERT2 /R26tdTom pups were injected with tamoxifen at P1 and perfused at 7 dpi. (Ai) Expected results with Notch1 receptor KO driving the progenitors towards the apical VSN fate. (B-Cii) Immunofluorescence anti Meis2, Tfap2e (AP-2epsilon) and tdTom in Ascl1Cre ERT2 induced control and Notch1 cKO pups at 14 dpi. Arrows highlight traced Meis2 + and arrowheads highlight Tfap2e + VSNs. (D,Di) Quantification of the percentage of traced VSNs that are Meis2 + , Tfap2e + , and Meis2/Tfap2e double negative cells in control and Notch1 KO mice at 7 dpi and 14 dpi. (E) Feature plots of Gnai2 ( Galphai2 ) and Gnao1 ( Galphao ) genes in the Seurat object 2 show that both G proteins are transcriptionally active in both VSN branches at the early stages of the dichotomy before getting restricted to apical versus basal VSN branches, respectively (see dotted circle). (F,G) Immunofluorescence of Gnao1 /tdTom in control and Notch1 cKOs at 14 dpi. Arrows indicate Gnao1 expression in traced cells. (H,I) Quantification of the percentage of traced VSNs that are Gnao1 + and Gnai2 + in control and Notch1 KO mice at 14 dpi. D, Di, H, and I, statistical analysis based on arcsine-transformed values of the percentage data; unpaired two-tailed t -test; n =3 biological replicates. Data shown as mean+-s.e.m. At 7 dpi, the average number of tdTom + cells in control group was 234.2
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunohistochemistry
Immunohistochemistry