Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Immunocytochemistry [2]
- Chromatin Immunoprecipitation [2]
- Other assay [3]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 710701 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- JunD Recombinant Superclonal™ Antibody (2HCLC)
- Antibody type
- Other
- Antigen
- Synthetic peptide
- Description
- This antibody is predicted to react with monkey, sheep, mouse, rat, zebra fish and bovine based on sequence homology. Recombinant rabbit Superclonal™ antibodies are unique offerings from Thermo Fisher Scientific. They are comprised of a selection of multiple different recombinant monoclonal antibodies, providing the best of both worlds - the sensitivity of polyclonal antibodies with the specificity of monoclonal antibodies - all delivered with the consistency only found in a recombinant antibody. While functionally the same as a polyclonal antibody - recognizing multiple epitope sites on the target and producing higher detection sensitivity for low abundance targets - a recombinant rabbit Superclonal™ antibody has a known mixture of light and heavy chains. The exact population can be produced in every lot, circumventing the biological variability typically associated with polyclonal antibody production. Note: Formerly called “Recombinant polyclonal antibody”, this product is now rebranded as “Recombinant Superclonal™ antibody”. The physical product and the performance remain unchanged.
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Antibody clone number
- 2HCLC
- Vial size
- 100 μg
- Concentration
- 0.5 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references Enhanced JunD/RSK3 signalling due to loss of BRD4/FOXD3/miR-548d-3p axis determines BET inhibition resistance.
Tai F, Gong K, Song K, He Y, Shi J
Nature communications 2020 Jan 14;11(1):258
Nature communications 2020 Jan 14;11(1):258
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
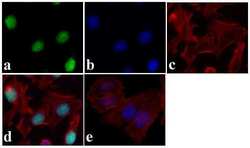
- Experimental details
- Immunofluorescent analysis of JunD was done on 70% confluent log phase HeLa cells. The cells were fixed with 4% paraformaldehyde for 15 minutes, permeabilized with 0.25% Triton X-100 for 10 minutes, and blocked with 5% BSA for 1 hour at room temperature. The cells were labeled with JunD Recombinant Rabbit Polyclonal Antibody (Product # 710701) at 5 µg/mL in 1% BSA and incubated for 3 hours at room temperature and then labeled with Alexa Fluor 488 Goat anti-Rabbit IgG Secondary Antibody (Product # A-11008) at a dilution of 1:400 for 30 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (Product # S36938). F-actin (Panel c: red) was stained with Alexa Fluor 594 Phalloidin (Product # A12381). Panel d is a merged image showing nuclear localization. Panel e shows no primary antibody control. The images were captured at 20X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
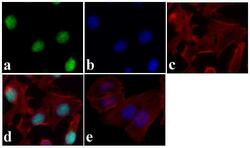
- Experimental details
- Immunofluorescent analysis of JunD was done on 70% confluent log phase HeLa cells. The cells were fixed with 4% paraformaldehyde for 15 minutes, permeabilized with 0.25% Triton X-100 for 10 minutes, and blocked with 5% BSA for 1 hour at room temperature. The cells were labeled with JunD Recombinant Rabbit Superclonal™ Antibody (Product # 710701) at 5 µg/mL in 1% BSA and incubated for 3 hours at room temperature and then labeled with Alexa Fluor 488 Goat anti-Rabbit IgG Secondary Antibody (Product # A-11008) at a dilution of 1:400 for 30 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (Product # S36938). F-actin (Panel c: red) was stained with Alexa Fluor 594 Phalloidin (Product # A12381). Panel d is a merged image showing nuclear localization. Panel e shows no primary antibody control. The images were captured at 20X magnification.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
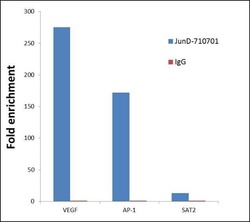
- Experimental details
- ChIP- qPCR analysis of JunD was performed with 5 µg of the Jun D Recombinant Rabbit Polyclonal Antibody (Product # 710701) on sheared chromatin from 2 million HeLa cells (serum starved for 24 hours and serum released for 2 hours) using the MAGnify Chromatin Immunoprecipitation System (Product # 49-2024). Normal Rabbit IgG was used as a negative IP control. The purified DNA from each ChIP sample was analyzed by StepOnePlus Real-Time PCR System (Product # 4376600) with primers for the promoter of active VEGF, AP-1 gene loci, used as positive control target gene, and the inactive SAT2 gene loci, used as negative control target gene. Data is presented as fold enrichment of the antibody signal versus the negative control IgG using the comparative CT method.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
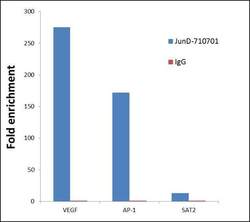
- Experimental details
- ChIP- qPCR analysis of JunD was performed with 5 µg of the Jun D Recombinant Rabbit Superclonal™ Antibody (Product # 710701) on sheared chromatin from 2 million HeLa cells (serum starved for 24 hours and serum released for 2 hours) using the MAGnify Chromatin Immunoprecipitation System (Product # 49-2024). Normal Rabbit IgG was used as a negative IP control. The purified DNA from each ChIP sample was analyzed by StepOnePlus Real-Time PCR System (Product # 4376600) with primers for the promoter of active VEGF, AP-1 gene loci, used as positive control target gene, and the inactive SAT2 gene loci, used as negative control target gene. Data is presented as fold enrichment of the antibody signal versus the negative control IgG using the comparative CT method.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
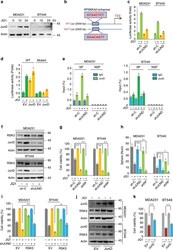
- Experimental details
- Fig. 2 JunD-dependent RPS6KA2 transcription mediates BETi resistance. a Western blotting was performed to detect JunD protein levels, MDA-MB-231 and BT549 cells were treated with DMSO or JQ1 (1 muM) for 0, 12 and 24 h. b Photograph depicted the potential JunD binding site in the enhancer region of RPS6KA2 gene. Wild-type and mutant RPS6KA2 gene enhancer luciferase plasmids are shown. c Luciferase assays were performed in MDA-MB-231 and BT549 cells transfected with shRNA for JunD and control, in the presence of DMSO or JQ1 (1 muM) for 6 h. Data are reported as mean +- SD. d Luciferase assays were performed in HEK293T cells transfected with wild-type or mutant RPS6KA2 enhancer luciferase construct with or without JUND co-transfection. HEK293T cells were treated with DMSO or JQ1 (1 muM) for 6 h. Data are reported as mean +- SD. e Chromatin immunoprecipitation (ChIP)-qPCR assay was executed in JUND -knockdown BLBC cells and their vector controls treated with DMSO or JQ1 (1 muM) for 6 h. 'SP' indicates specific primers of ChIP for RPS6KA2 gene enhancer and 'NSP' indicates non-specific primers that recognize the region downstream of the 3' end of the gene. f Western blotting was performed to detect the expression levels of JunD and RSK3 in control and JUND -knockdown clones of MDA-MB-231 and BT549. g JUND or RPS6KA2-knockdown MDA-MB-231 and BT549 cells as well as their vector control cells were treated with DMSO or JQ1 (1 muM) for 48 h, and luminescent cell viability assays were do
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
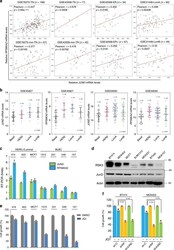
- Experimental details
- Fig. 3 JunD/RSK3 signalling correlates to BET inhibition sensitivity. a Correlation analyses of JUND and RPS6KA2 in four datasets of breast cancer patients from GEO. GSE76275 was separated into two sub-groups: triple-negative (TN) and non-triple-negative (non-TN); GSE43358 was separated into two sub-groups: triple-negative (TN) and non-triple-negative (non-TN); GSE42568 was separated into two sub-groups: ER-positive (ER+) and ER-negative (ER-); GSE31448 was separated into two sub-groups: luminal A (lumA) and luminal B (lumB). Pearson Coefficients of correlation and p values are shown. b Gene differential expression analyses of the mRNA levels of JUND and RPS6KA2 were conducted in two datasets that contained four subtypes of breast cancer patients, including BLBC, luminal A, luminal B and HER2 + . c The mRNA expression statuses of JUND and RPS6KA2 were detected in a panel of breast cancer cell lines by quantitative PCR assay. d Protein expression levels of JunD and RSK3 were detected in a panel of breast cancer cell lines by western blotting. e Luminal and HER2 + breast cancer cell lines as well as BLBC cell lines were treated with DMSO or JQ1 (1 muM) for 48 h, cell growth was detected by CCK8 assay. Statistical data (mean +- SD) are shown. f RPS6KA2 and JUND genes were knocked down in two JQ1-resistant breast cancer cell lines, cell growth was measured by CCK8 assay. Statistical data (mean +- SD) are shown (*** P < 0.001, one-way ANOVA). Source data are provided as a Source D
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
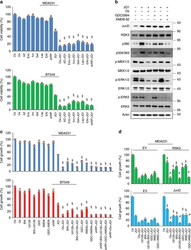
- Experimental details
- Fig. 5 Targeting EGFR/MEKs/ERKs reverses BETi resistance. a Measurement of cell viability of MDA-MB231 and BT549 cells by CellTiter-Glo (r) luminescent viability assay. Control and RPS6KA2 -knockdown cells were treated with JQ1 in the absence or presence of seven EGFR inhibitors for 48 h. 1: Ctr; 2: osimertinib; 3: AZ5104; 4: erlotinib; 5: icotinib; 6: gefitinib; 7: lapatinib; 8: EAI045; 9: RPS6KA2 -shRNA; 10: JQ1; 11: JQ1 + osimertinib; 12: JQ1 + AZ5104; 13: JQ1 + erlotinib; 14: JQ1 + icotinib; 15: JQ1 + gefitinib; 16: JQ1 + lapatinib; 17: JQ1 + EAI045; 18: JQ1 + RPS6KA2 -shRNA. Statistical data (Mean +- SD) are shown. P values were calculated between single JQ1-treated samples and samples treated with JQ1 plus EGFR inhibitors. Symbol 'SS' indicates statistical significance ( P < 0.05, one-way ANOVA). b MDA-MB-231 cells were treated with JQ1 (1 muM) and/or osimertinib, GDC0994, XMD8-92 (1 muM) or GDC0994/XMD8-92 for 12 h, western blotting was performed to detect JunD protein expression, and phosphorylated and total levels of MEK1/2, ERK1/2, ERK5 and RSK3. c CCK8 assays were done to measure the killing effects. Control and RPS6KA2 -knockdown MDA-MB-231 and BT549 cells were treated with indicated the inhibitors for 48 h. 1: Control; 2: osimertinib; 3: BIX-02188; 4: U0126; 5: U0126 + BIX-02188; 6: GDC0994; 7: XMD8-92; 8: GDC0994 + XMD8-92; 9: RPS6KA2 -shRNA; 10: JQ1; 11: JQ1 + osimertinib; 12: JQ1 + BIX-02188; 13: JQ1 + U0126; 14: JQ1 + BIX-02188 + U0126; 15: JQ1 + GDC0994; 16:
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry