MA5-13449
antibody from Invitrogen Antibodies
Targeting: SNRPB
COD, Sm-B/B', SmB/SmB', snRNP-B, SNRPB1
Antibody data
- Antibody Data
- Antigen structure
- References [29]
- Comments [0]
- Validations
- Immunocytochemistry [4]
- Immunohistochemistry [1]
- Other assay [7]
Submit
Validation data
Reference
Comment
Report error
- Product number
- MA5-13449 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- SNRPB Monoclonal Antibody (Y12)
- Antibody type
- Monoclonal
- Antigen
- Other
- Reactivity
- Human, Mouse, Rat, Drosophila, Porcine, Rabbit
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- Y12
- Vial size
- 500 μL
- Concentration
- 200 μg/mL
- Storage
- 4°C
Submitted references The integrity of the U12 snRNA 3' stem-loop is necessary for its overall stability.
Therapeutic manipulation of IKBKAP mis-splicing with a small molecule to cure familial dysautonomia.
Tau-Mediated Disruption of the Spliceosome Triggers Cryptic RNA Splicing and Neurodegeneration in Alzheimer's Disease.
PRMT1 loss sensitizes cells to PRMT5 inhibition.
Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa.
Checkpoints for Autoreactive B Cells in the Peripheral Blood of Lupus Patients Assessed by Flow Cytometry.
An Animal Model Using Metallic Ions to Produce Autoimmune Nephritis.
In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea.
Spliceosome integrity is defective in the motor neuron diseases ALS and SMA.
Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration.
Combination of SMN trans-splicing and a neurotrophic factor increases the life span and body mass in a severe model of spinal muscular atrophy.
The inability of fully grown germinal vesicle stage oocyte cytoplasm to transcriptionally silence transferred transcribing nuclei.
Functional association of the Microprocessor complex with the spliceosome.
Conserved patterns of nuclear compartmentalization are not observed in the chordate Oikopleura.
Inhibition of U snRNP assembly by a virus-encoded proteinase.
Mutations in splicing factor PRPF3, causing retinal degeneration, form detrimental aggregates in photoreceptor cells.
Myosin16b: The COOH-tail region directs localization to the nucleus and overexpression delays S-phase progression.
Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila.
Restoration of SMN function: delivery of a trans-splicing RNA re-directs SMN2 pre-mRNA splicing.
Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly.
Gemin8 is required for the architecture and function of the survival motor neuron complex.
Nuclear substructure reorganization during late-stage erythropoiesis is selective and does not involve caspase cleavage of major nuclear substructural proteins.
Subcellular localization of protein kinase C delta and epsilon affects transcriptional and post-transcriptional processes in four-cell mouse embryos.
Tim50a, a nuclear isoform of the mitochondrial Tim50, interacts with proteins involved in snRNP biogenesis.
A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules.
Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein.
ELL and EAF1 are Cajal body components that are disrupted in MLL-ELL leukemia.
pICln inhibits snRNP biogenesis by binding core spliceosomal proteins.
pICln inhibits snRNP biogenesis by binding core spliceosomal proteins.
Norppa AJ, Frilander MJ
Nucleic acids research 2021 Mar 18;49(5):2835-2847
Nucleic acids research 2021 Mar 18;49(5):2835-2847
Therapeutic manipulation of IKBKAP mis-splicing with a small molecule to cure familial dysautonomia.
Ajiro M, Awaya T, Kim YJ, Iida K, Denawa M, Tanaka N, Kurosawa R, Matsushima S, Shibata S, Sakamoto T, Studer L, Krainer AR, Hagiwara M
Nature communications 2021 Jul 23;12(1):4507
Nature communications 2021 Jul 23;12(1):4507
Tau-Mediated Disruption of the Spliceosome Triggers Cryptic RNA Splicing and Neurodegeneration in Alzheimer's Disease.
Hsieh YC, Guo C, Yalamanchili HK, Abreha M, Al-Ouran R, Li Y, Dammer EB, Lah JJ, Levey AI, Bennett DA, De Jager PL, Seyfried NT, Liu Z, Shulman JM
Cell reports 2019 Oct 8;29(2):301-316.e10
Cell reports 2019 Oct 8;29(2):301-316.e10
PRMT1 loss sensitizes cells to PRMT5 inhibition.
Gao G, Zhang L, Villarreal OD, He W, Su D, Bedford E, Moh P, Shen J, Shi X, Bedford MT, Xu H
Nucleic acids research 2019 Jun 4;47(10):5038-5048
Nucleic acids research 2019 Jun 4;47(10):5038-5048
Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa.
Buskin A, Zhu L, Chichagova V, Basu B, Mozaffari-Jovin S, Dolan D, Droop A, Collin J, Bronstein R, Mehrotra S, Farkas M, Hilgen G, White K, Pan KT, Treumann A, Hallam D, Bialas K, Chung G, Mellough C, Ding Y, Krasnogor N, Przyborski S, Zwolinski S, Al-Aama J, Alharthi S, Xu Y, Wheway G, Szymanska K, McKibbin M, Inglehearn CF, Elliott DJ, Lindsay S, Ali RR, Steel DH, Armstrong L, Sernagor E, Urlaub H, Pierce E, Lührmann R, Grellscheid SN, Johnson CA, Lako M
Nature communications 2018 Oct 12;9(1):4234
Nature communications 2018 Oct 12;9(1):4234
Checkpoints for Autoreactive B Cells in the Peripheral Blood of Lupus Patients Assessed by Flow Cytometry.
Malkiel S, Jeganathan V, Wolfson S, Manjarrez Orduño N, Marasco E, Aranow C, Mackay M, Gregersen PK, Diamond B
Arthritis & rheumatology (Hoboken, N.J.) 2016 Sep;68(9):2210-20
Arthritis & rheumatology (Hoboken, N.J.) 2016 Sep;68(9):2210-20
An Animal Model Using Metallic Ions to Produce Autoimmune Nephritis.
Ramírez-Sandoval R, Luévano-Rodríguez N, Rodríguez-Rodríguez M, Pérez-Pérez ME, Saldívar-Elias S, Gurrola-Carlos R, Avalos-Díaz E, Bollain-y-Goytia JJ, Herrera-Esparza R
Journal of immunology research 2015;2015:269610
Journal of immunology research 2015;2015:269610
In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea.
King RS, Newmark PA
BMC developmental biology 2013 Mar 12;13:8
BMC developmental biology 2013 Mar 12;13:8
Spliceosome integrity is defective in the motor neuron diseases ALS and SMA.
Tsuiji H, Iguchi Y, Furuya A, Kataoka A, Hatsuta H, Atsuta N, Tanaka F, Hashizume Y, Akatsu H, Murayama S, Sobue G, Yamanaka K
EMBO molecular medicine 2013 Feb;5(2):221-34
EMBO molecular medicine 2013 Feb;5(2):221-34
Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration.
Jia Y, Mu JC, Ackerman SL
Cell 2012 Jan 20;148(1-2):296-308
Cell 2012 Jan 20;148(1-2):296-308
Combination of SMN trans-splicing and a neurotrophic factor increases the life span and body mass in a severe model of spinal muscular atrophy.
Shababi M, Glascock J, Lorson CL
Human gene therapy 2011 Feb;22(2):135-44
Human gene therapy 2011 Feb;22(2):135-44
The inability of fully grown germinal vesicle stage oocyte cytoplasm to transcriptionally silence transferred transcribing nuclei.
Fulka H, Novakova Z, Mosko T, Fulka J Jr
Histochemistry and cell biology 2009 Oct;132(4):457-68
Histochemistry and cell biology 2009 Oct;132(4):457-68
Functional association of the Microprocessor complex with the spliceosome.
Kataoka N, Fujita M, Ohno M
Molecular and cellular biology 2009 Jun;29(12):3243-54
Molecular and cellular biology 2009 Jun;29(12):3243-54
Conserved patterns of nuclear compartmentalization are not observed in the chordate Oikopleura.
Spada F, Koch J, Sadoni N, Mitchell N, Ganot P, De Boni U, Zink D, Thompson EM
Biology of the cell 2007 May;99(5):273-87
Biology of the cell 2007 May;99(5):273-87
Inhibition of U snRNP assembly by a virus-encoded proteinase.
Almstead LL, Sarnow P
Genes & development 2007 May 1;21(9):1086-97
Genes & development 2007 May 1;21(9):1086-97
Mutations in splicing factor PRPF3, causing retinal degeneration, form detrimental aggregates in photoreceptor cells.
Comitato A, Spampanato C, Chakarova C, Sanges D, Bhattacharya SS, Marigo V
Human molecular genetics 2007 Jul 15;16(14):1699-707
Human molecular genetics 2007 Jul 15;16(14):1699-707
Myosin16b: The COOH-tail region directs localization to the nucleus and overexpression delays S-phase progression.
Cameron RS, Liu C, Mixon AS, Pihkala JP, Rahn RJ, Cameron PL
Cell motility and the cytoskeleton 2007 Jan;64(1):19-48
Cell motility and the cytoskeleton 2007 Jan;64(1):19-48
Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila.
Anne J, Ollo R, Ephrussi A, Mechler BM
Development (Cambridge, England) 2007 Jan;134(1):137-46
Development (Cambridge, England) 2007 Jan;134(1):137-46
Restoration of SMN function: delivery of a trans-splicing RNA re-directs SMN2 pre-mRNA splicing.
Coady TH, Shababi M, Tullis GE, Lorson CL
Molecular therapy : the journal of the American Society of Gene Therapy 2007 Aug;15(8):1471-8
Molecular therapy : the journal of the American Society of Gene Therapy 2007 Aug;15(8):1471-8
Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly.
Carissimi C, Saieva L, Baccon J, Chiarella P, Maiolica A, Sawyer A, Rappsilber J, Pellizzoni L
The Journal of biological chemistry 2006 Mar 24;281(12):8126-34
The Journal of biological chemistry 2006 Mar 24;281(12):8126-34
Gemin8 is required for the architecture and function of the survival motor neuron complex.
Carissimi C, Saieva L, Gabanella F, Pellizzoni L
The Journal of biological chemistry 2006 Dec 1;281(48):37009-16
The Journal of biological chemistry 2006 Dec 1;281(48):37009-16
Nuclear substructure reorganization during late-stage erythropoiesis is selective and does not involve caspase cleavage of major nuclear substructural proteins.
Krauss SW, Lo AJ, Short SA, Koury MJ, Mohandas N, Chasis JA
Blood 2005 Sep 15;106(6):2200-5
Blood 2005 Sep 15;106(6):2200-5
Subcellular localization of protein kinase C delta and epsilon affects transcriptional and post-transcriptional processes in four-cell mouse embryos.
Dehghani H, Reith C, Hahnel AC
Reproduction (Cambridge, England) 2005 Oct;130(4):453-65
Reproduction (Cambridge, England) 2005 Oct;130(4):453-65
Tim50a, a nuclear isoform of the mitochondrial Tim50, interacts with proteins involved in snRNP biogenesis.
Xu H, Somers ZB, Robinson ML 2nd, Hebert MD
BMC cell biology 2005 Jul 11;6(1):29
BMC cell biology 2005 Jul 11;6(1):29
A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules.
Metz A, Soret J, Vourc'h C, Tazi J, Jolly C
Journal of cell science 2004 Sep 1;117(Pt 19):4551-8
Journal of cell science 2004 Sep 1;117(Pt 19):4551-8
Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein.
Narayanan U, Achsel T, Lührmann R, Matera AG
Molecular cell 2004 Oct 22;16(2):223-34
Molecular cell 2004 Oct 22;16(2):223-34
ELL and EAF1 are Cajal body components that are disrupted in MLL-ELL leukemia.
Polak PE, Simone F, Kaberlein JJ, Luo RT, Thirman MJ
Molecular biology of the cell 2003 Apr;14(4):1517-28
Molecular biology of the cell 2003 Apr;14(4):1517-28
pICln inhibits snRNP biogenesis by binding core spliceosomal proteins.
Pu WT, Krapivinsky GB, Krapivinsky L, Clapham DE
Molecular and cellular biology 1999 Jun;19(6):4113-20
Molecular and cellular biology 1999 Jun;19(6):4113-20
pICln inhibits snRNP biogenesis by binding core spliceosomal proteins.
Pu WT, Krapivinsky GB, Krapivinsky L, Clapham DE
Molecular and cellular biology 1999 Jun;19(6):4113-20
Molecular and cellular biology 1999 Jun;19(6):4113-20
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
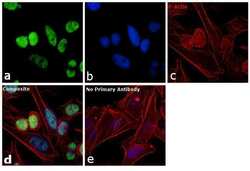
- Experimental details
- Immunofluorescence analysis of SNRPB was performed using 70% confluent log phase HeLa cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 10 minutes, and blocked with 1% BSA for 1 hour at room temperature. The cells were labeled with SNRPB Mouse monoclonal antibody (Product # MA5-13449) at 2 µg/mL in 0.1% BSA and incubated for 3 hours at room temperature and then labeled with Goat anti-Mouse IgG (H+L) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate (Product # A28175) at a dilution of 1:2000 for 45 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (S36938). F-actin (Panel c: red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image showing nuclear localization. Panel e shows the no primary antibody control. The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
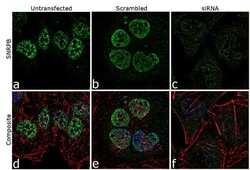
- Experimental details
- Knockdown of SNRPB was achieved by transfecting HeLa cells with SNRPB specific siRNA (Silencer® select Cat # s13220, s13221). Immunofluorescence analysis was performed on HeLa cells (untransfected, panel a,d), transfected with non-specific scrambled siRNA (panels b,e) and transfected with SNRPB specific siRNA (panel c,f) Cells were fixed, permeabilized, and labelled with SNRPB Mouse monoclonal Antibody (Product # MA5-13449, 5 µg/mL), followed by Goat anti-Mouse IgG (H+L) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate (Product # A28175, 1:2000). Nuclei (blue) were stained using SlowFade® Gold Antifade Mountant with DAPI (Product # S36938), and Rhodamine Phalloidin (Product # R415, 1:300) was used for cytoskeletal F-actin (red) staining. Reduction of specific signal was observed upon siRNA mediated knockdown (panel c,f) confirming specificity of the antibody to SNRPB(green). The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
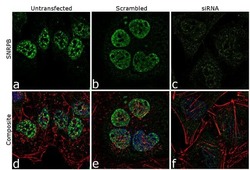
- Experimental details
- Knockdown of SNRPB was achieved by transfecting HeLa cells with SNRPB specific siRNA (Silencer® select Cat # s13220, s13221). Immunofluorescence analysis was performed on HeLa cells (untransfected, panel a,d), transfected with non-specific scrambled siRNA (panels b,e) and transfected with SNRPB specific siRNA (panel c,f) Cells were fixed, permeabilized, and labelled with SNRPB Mouse monoclonal Antibody (Product # MA5-13449, 5 µg/mL), followed by Goat anti-Mouse IgG (H+L) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate (Product # A28175, 1:2000). Nuclei (blue) were stained using SlowFade® Gold Antifade Mountant with DAPI (Product # S36938), and Rhodamine Phalloidin (Product # R415, 1:300) was used for cytoskeletal F-actin (red) staining. Reduction of specific signal was observed upon siRNA mediated knockdown (panel c,f) confirming specificity of the antibody to SNRPB(green). The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
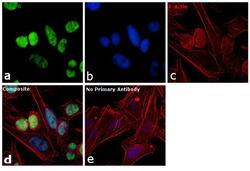
- Experimental details
- Immunofluorescence analysis of SNRPB was performed using 70% confluent log phase HeLa cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 10 minutes, and blocked with 1% BSA for 1 hour at room temperature. The cells were labeled with SNRPB Mouse monoclonal antibody (Product # MA5-13449) at 2 µg/mL in 0.1% BSA and incubated for 3 hours at room temperature and then labeled with Goat anti-Mouse IgG (H+L) Superclonal™ Secondary Antibody, Alexa Fluor® 488 conjugate (Product # A28175) at a dilution of 1:2000 for 45 minutes at room temperature (Panel a: green). Nuclei (Panel b: blue) were stained with SlowFade® Gold Antifade Mountant with DAPI (S36938). F-actin (Panel c: red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image showing nuclear localization. Panel e shows the no primary antibody control. The images were captured at 60X magnification.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
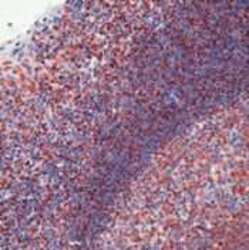
- Experimental details
- Formalin-fixed, paraffin-embedded human tonsil stained with sm antibody using peroxidase-conjugate and AEC chromogen. Note nuclear staining of cells.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
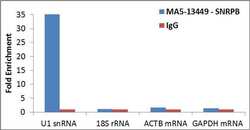
- Experimental details
- Detection of binding of endogenous SNRPB protein to specific RNA using Anti-SNRPB Antibody: RNA Immunoprecipitation (RIP) was performed using Anti-SNRPB Mouse Monoclonal Antibody (Product # MA5-13449, 3 µg) on clarified whole cell lysate from 4 million HCT 116 cells. Normal Rabbit IgG was used as a negative IP control. Immunoprecipitated RNA was purified by RiboPure™ RNA Purification Kit (Product # AM1924) and analyzed by RT-PCR using the Power SYBR® Green RNA-to-CT™ 1-Step Kit (Product # 4389986) with primer pairs over U1 snRNA (positive) and 18S rRNA, ACTB mRNA, GAPDH mRNA (negative). Data is presented as fold enrichment of the antibody signal versus the negative control IgG using the comparative CT method.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
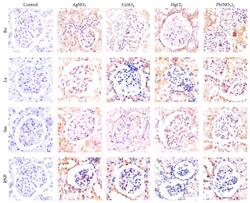
- Experimental details
- Figure 5 Immunohistochemistry of kidneys demonstrating the overexpression of ribonucleoproteins induced by heavy metal salts. Note that at the beginning of the trial (control), the expression is faint or absent, whereas at the end of the intoxication trial, ribonucleoprotein expression notably increases.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
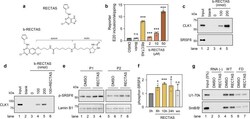
- Experimental details
- Fig. 3 RECTAS directly interacts with CDC-like kinase 1 (CLK1), and it enhances cellular SRSF6 activity. a Structures of RECTAS and b-RECTAS. b Extent of IKBKAP -FD exon 20 inclusion represented by GFP/RFP ratio, following treatment with biotin (50 uM), RECTAS (2 uM), b-RECTAS (2, 10, and 50 uM), or solvent only (0.1% DMSO) for 24 h in reporter-transfected HeLa cells. c , d Western blotting of pull-down products by b-RECTAS (0, 100, or 200 nmol/reaction) for HeLa cells transfected with a flag-tagged CLK1 expression vector ( c ) or GST-tagged recombinant CLK1 ( d ). RECTAS (500 nmol) was incubated with 100 nmol b-RECTAS for the competition assay in ( d ). e , f Western blot ( e ) for phosphor SRSF6 in FD patient primary fibroblasts (P1 and P2), treated with RECTAS (10 uM) or solvent only (0.1% DMSO) for 24 h. f Phosphor SRSF6 in FD fibroblasts (P1) was quantified at 0, 6, 10, 24 h after RECTAS (10 uM) treatment (columns, 0 h, 6 h, 10 h, and 24 h), as well as 10 h after washout of compound following RECTAS treatment (10 uM) for 24 h (column, wo). Lamin B1 served as a loading control in ( e ). g Western blot for U1-70k and SmB/B' in pull-down products with biotin-conjugated RNA surrounding IKBKAP exon 20 donor site with (oAM154) or without (oAM153) IVS20 + 6 T > C mutation (labeled as FD and WT, respectively). Pull-down was conducted in the presence of 10 uM RECTAS with 1% DMSO, or 1% DMSO only. RNA (-), control sample without RNA bait. Input, HeLa nuclear extract of 5% input am
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
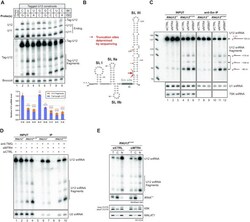
- Figure 3. Identification and characterization of truncated U12 snRNAs in human cells. ( A ) Effects of mutations in the terminal base-pair of U12 snRNA stem-loop III on full-length U12 and U12 fragment levels. Top: Plasmid constructs for expressing tag-U12 with various combinations of nucleotides in the 84 and 150 positions were transfected into HeLa cells. Co-transfected F30-2xdBroccoli expression plasmid served as a transfection control. Total RNA from transfected HeLa cells was analyzed by northern blot with U12, U11, tag-U12 and Broccoli probes. Bottom: Quantification of full-length U12 (blue) and U12 fragment (orange) levels. Band intensities were normalized by the sum of all band intensities of each replicate, and the mean full-length U12 snRNA signal of WT tag-U12 (C-G) was then set to 1. Statistical significance was determined using one-way ANOVA followed by Dunnett's test. Significance levels above each bar refer to comparisons between WT tag-U12 and the indicated variant. Based on pair-wise comparisons (ANOVA with Tukey's test), differences in fragment levels between the group of variants showing low levels of tag-U12 fragments (C-G, G-C, G-G) and the group with high levels of fragments (U-G, U-A, C-C, U-C, C-A) were all statistically significant (*-***). In contrast, none of the pair-wise comparisons within the two groups showed statistically significant differences in fragment levels. ( B ) Location of truncation sites of U12 snRNA fragments determined by Sanger s
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
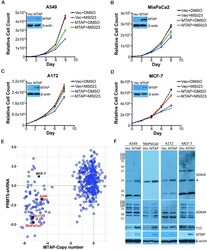
- Figure 4. Different sensitivities of MTAP -deleted and MTAP-rescued cells to Type I PRMT inhibition. ( A-D ) MTAP -deleted cells were reconstituted with the stable re-expression of MTAP. These isogenic cell pairs were cultured for 8 days in the presence of DMSO or 1 muM of MS023 (0.1 muM for MiaPaCa2 cells). Different cell types display differing degrees of sensitivity to MS023, which we established empirically. The cell growth curves are plotted for A549 (A), MiaPaCa2 (B), A172 (C) and MCF-7 (D) cell lines. The expression levels of MTAP are shown for each pair of cell lines. ( E ) A scatter plot showing the sensitivities of MTAP -deleted cells to PRMT5 knockdown. Each dot in the scatter plot represents a cancer cell line. Data retrieved from the DepMap project ( https://depmap.org/portal/ ). ( F ) Using total cell lysates from MTAP -deleted and MTAP-rescued cells, the levels of SDMA, ADMA, SmB methylation (Y12) and MTAP were analyzed by western blot.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
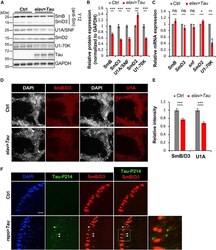
- Figure 2. Disruption of the Spliceosome following Tau Expression in Drosophila (A and B) Pan-neuronal human Tau R406W disrupts expression of multiple spliceosomal proteins in 1-day-old adult fly head homogenates. Western blots (A) were probed for Tau or spliceosome proteins and normalized to the loading control, GAPDH (n = 4 replicates for quantification, B). The Y12 antibody recognizes both SmB and SmD3. (C) mRNA expression was also examined in 1-day-old adult heads (n = 3). (D and E) Whole-mount stains of 1-day-old adult fly brains (D) reveal depletion of SmB/D3 and U1A/SNF protein (red, n = 15 for quantification, E). Nuclei are colabeled with 4',6-diamidino-2-phenylindole (DAPI; grayscale). Scale bar: 20 mum. (F) Glial expression of Tau WT ( repo-GAL4 ) induces cytoplasmic foci (arrowheads) of SmB/D3 (Y12, red) that colocalize with phospho-Tau aggregates (green) in 10-day-old adult brains. Nuclei are labeled with DAPI (blue). Boxed region is magnified at right. Quantification (n > 9) reveals 14.86% +- 2.3% of phospho-Tau aggregates colabeling for Sm proteins; aggregates were not observed in controls. Scale bar: 10 mum. See also Figure S3A . All error bars denote mean +- SEM. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
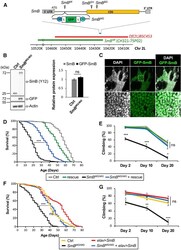
- Figure 3. Loss of SmB , Encoding a Core Spliceosome Factor, Causes Reduced Survival and Progressive Locomotor Impairment (A) The SmB locus, including single coding exon (CDS, yellow), untranslated regions (UTRs, gray), and transposable elements. To generate SmB MG , recombination-mediated cassette exchange was performed using SmB MI , introducing a coding exon for GFP, flanked by flexible linkers (Ls) and splice acceptor and donor sequences (SA/SDs). The deficiency strain (red), Df(2L)BSC453 , deletes ~51 kb including the entire SmB locus. The transgenic genomic rescue strain, SmB GR , carries a ~90-kb bacterial artificial chromosome (BAC, CH321-75P02, green), including SmB. See also Table S2 . (B) SmB MG/MG homozygotes demonstrate expression of the GFP-SmB fusion protein at levels comparable with controls (n = 3 for quantification, SmB protein normalized to Actin). (C) GFP-SmB (green) is localized to the nucleus (DAPI, grayscale) in brains from SmB MG/MG adults. Scale bar: 20 mum. Boxed region (top) is magnified below. (D) SmB MG/MG adults (black) exhibit reduced survival and this phenotype is partially rescued by the SmB genomic rescue (blue) (n > 313 per genotype). See also Figure S4A . (E) SmB MG/MG adults also manifest progressive locomotor impairment (n > 5 groups). See also Figure S4B . (F and G) Both the SmB MG/MG survival (F, n > 288) and locomotor (G, n > 4 groups) phenotypes were rescued by pan-neuronal expression of wild-type SmB . All error bars denote mean +- SE
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry Immunoelectron microscopy
Immunoelectron microscopy