PA5-27195
antibody from Invitrogen Antibodies
Targeting: RAD51
BRCC5, FANCR, HsRad51, HsT16930, RAD51A, RECA
Antibody data
- Antibody Data
- Antigen structure
- References [12]
- Comments [0]
- Validations
- Immunocytochemistry [2]
- Immunoprecipitation [1]
- Immunohistochemistry [3]
- Other assay [10]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-27195 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- RAD51 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant full-length protein
- Description
- Recommended positive controls: 293T, A431, HeLa, HepG2, Neuro2A, C8D30, NIH-3T3, Raw264.7, C2C12, PC-12, Rat-2. Predicted reactivity: Mouse (98%), Rat (98%), Zebrafish (90%), Xenopus laevis (96%), Dog (98%), Pig (99%), Rabbit (97%), Chicken (95%), Bovine (98%). Store product as a concentrated solution. Centrifuge briefly prior to opening the vial.
- Reactivity
- Human, Mouse, Rat, Zebrafish
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 0.14 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references RAD51 is essential for spermatogenesis and male fertility in mice.
Selinexor is a novel inhibitor of DNA damage response in Merkel cell carcinoma.
Gut Microbiota Dysbiosis Accelerates Prostate Cancer Progression Through Increased LPCAT1 Expression and Enhanced DNA Repair Pathways.
CHK1 protects oncogenic KRAS-expressing cells from DNA damage and is a target for pancreatic cancer treatment.
Chloroxine overrides DNA damage tolerance to restore platinum sensitivity in high-grade serous ovarian cancer.
The histone modification reader ZCWPW1 links histone methylation to PRDM9-induced double-strand break repair.
MEIOK21: a new component of meiotic recombination bridges required for spermatogenesis.
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells.
SCRE serves as a unique synaptonemal complex fastener and is essential for progression of meiosis prophase I in mice.
A meiosis-specific BRCA2 binding protein recruits recombinases to DNA double-strand breaks to ensure homologous recombination.
Drugging DNA repair to target T-ALL cells.
Rgf1p (Rho1p GEF) is required for double-strand break repair in fission yeast.
Qin J, Huang T, Wang J, Xu L, Dang Q, Xu X, Liu H, Liu Z, Shao C, Zhang X
Cell death discovery 2022 Mar 15;8(1):118
Cell death discovery 2022 Mar 15;8(1):118
Selinexor is a novel inhibitor of DNA damage response in Merkel cell carcinoma.
Moore SA, Narayanan D, Simonette RA, Bartley BR, Doan HQ, Rady PL, Tyring SK
Clinical and experimental dermatology 2022 Jul;47(7):1354-1357
Clinical and experimental dermatology 2022 Jul;47(7):1354-1357
Gut Microbiota Dysbiosis Accelerates Prostate Cancer Progression Through Increased LPCAT1 Expression and Enhanced DNA Repair Pathways.
Liu Y, Yang C, Zhang Z, Jiang H
Frontiers in oncology 2021;11:679712
Frontiers in oncology 2021;11:679712
CHK1 protects oncogenic KRAS-expressing cells from DNA damage and is a target for pancreatic cancer treatment.
Klomp JE, Lee YS, Goodwin CM, Papke B, Klomp JA, Waters AM, Stalnecker CA, DeLiberty JM, Drizyte-Miller K, Yang R, Diehl JN, Yin HH, Pierobon M, Baldelli E, Ryan MB, Li S, Peterson J, Smith AR, Neal JT, McCormick AK, Kuo CJ, Counter CM, Petricoin EF 3rd, Cox AD, Bryant KL, Der CJ
Cell reports 2021 Nov 30;37(9):110060
Cell reports 2021 Nov 30;37(9):110060
Chloroxine overrides DNA damage tolerance to restore platinum sensitivity in high-grade serous ovarian cancer.
Silva VL, Saxena J, Nicolini F, Hoare JI, Metcalf S, Martin SA, Lockley M
Cell death & disease 2021 Apr 14;12(4):395
Cell death & disease 2021 Apr 14;12(4):395
The histone modification reader ZCWPW1 links histone methylation to PRDM9-induced double-strand break repair.
Huang T, Yuan S, Gao L, Li M, Yu X, Zhan J, Yin Y, Liu C, Zhang C, Lu G, Li W, Liu J, Chen ZJ, Liu H
eLife 2020 May 6;9
eLife 2020 May 6;9
MEIOK21: a new component of meiotic recombination bridges required for spermatogenesis.
Shang Y, Huang T, Liu H, Liu Y, Liang H, Yu X, Li M, Zhai B, Yang X, Wei Y, Wang G, Chen Z, Wang S, Zhang L
Nucleic acids research 2020 Jul 9;48(12):6624-6639
Nucleic acids research 2020 Jul 9;48(12):6624-6639
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells.
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury DF, Alam I, Livera G, Emmanuelle M, Wang PJ, Nandakumar J, Davies OR, Shibuya H
Nature communications 2020 Apr 28;11(1):2055
Nature communications 2020 Apr 28;11(1):2055
SCRE serves as a unique synaptonemal complex fastener and is essential for progression of meiosis prophase I in mice.
Liu H, Huang T, Li M, Li M, Zhang C, Jiang J, Yu X, Yin Y, Zhang F, Lu G, Luo MC, Zhang LR, Li J, Liu K, Chen ZJ
Nucleic acids research 2019 Jun 20;47(11):5670-5683
Nucleic acids research 2019 Jun 20;47(11):5670-5683
A meiosis-specific BRCA2 binding protein recruits recombinases to DNA double-strand breaks to ensure homologous recombination.
Zhang J, Fujiwara Y, Yamamoto S, Shibuya H
Nature communications 2019 Feb 13;10(1):722
Nature communications 2019 Feb 13;10(1):722
Drugging DNA repair to target T-ALL cells.
Dasgupta Y, Golovine K, Nieborowska-Skorska M, Luo L, Matlawska-Wasowska K, Mullighan CG, Skorski T
Leukemia & lymphoma 2018 Jul;59(7):1746-1749
Leukemia & lymphoma 2018 Jul;59(7):1746-1749
Rgf1p (Rho1p GEF) is required for double-strand break repair in fission yeast.
Manjón E, Edreira T, Muñoz S, Sánchez Y
Nucleic acids research 2017 May 19;45(9):5269-5284
Nucleic acids research 2017 May 19;45(9):5269-5284
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
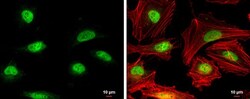
- Experimental details
- Immunocytochemistry-Immunofluorescence analysis of RAD51 was performed in HeLa cells fixed in 4% paraformaldehyde at RT for 15 min. Green: RAD51 Polyclonal Antibody (Product # PA5-27195) diluted at 1:500. Red: phalloidin, a cytoskeleton marker. Scale bar = 10 µm.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
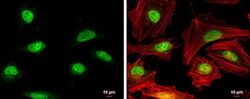
- Experimental details
- Immunocytochemistry-Immunofluorescence analysis of RAD51 was performed in HeLa cells fixed in 4% paraformaldehyde at RT for 15 min. Green: RAD51 Polyclonal Antibody (Product # PA5-27195) diluted at 1:500. Red: phalloidin, a cytoskeleton marker. Scale bar = 10 µm.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
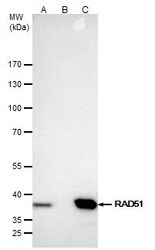
- Experimental details
- RAD51 Polyclonal Antibody immunoprecipitates Rad51 protein in IP experiments. IP samples: Jurkat whole cell extract. A. 40 µg Jurkat whole cell extract. B. Control with 4 µg of preimmune Rabbit IgG. C. Immunoprecipitation of Rad51 protein by 4 µg RAD51 Polyclonal Antibody (Product # PA5-27195). 5 % SDS-PAGE. The immunoprecipitated Rad51 protein was detected by RAD51 Polyclonal Antibody (Product # PA5-27195) diluted at 1:500.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
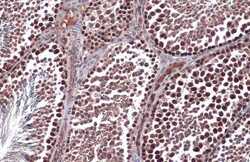
- Experimental details
- Immunohistochemistry (Paraffin) analysis of RAD51 was performed in paraffin-embedded mouse testis tissue using RAD51 Polyclonal Antibody (Product # PA5-27195) at a dilution of 1:500. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
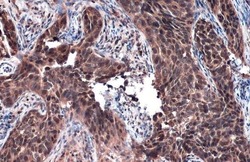
- Experimental details
- Immunohistochemistry (Paraffin) analysis of RAD51 was performed in paraffin-embedded human cervical carcinoma tissue using RAD51 Polyclonal Antibody (Product # PA5-27195) at a dilution of 1:500. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
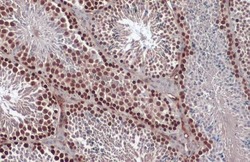
- Experimental details
- Immunohistochemistry (Paraffin) analysis of RAD51 was performed in paraffin-embedded mouse testis tissue using RAD51 Polyclonal Antibody (Product # PA5-27195) at a dilution of 1:1000. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
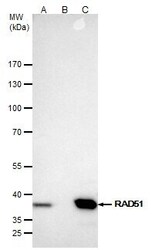
- RAD51 Polyclonal Antibody immunoprecipitates Rad51 protein in IP experiments. IP samples: Jurkat whole cell extract. A. 40 µg Jurkat whole cell extract. B. Control with 4 µg of preimmune Rabbit IgG. C. Immunoprecipitation of Rad51 protein by 4 µg RAD51 Polyclonal Antibody (Product # PA5-27195). 5 % SDS-PAGE. The immunoprecipitated Rad51 protein was detected by RAD51 Polyclonal Antibody (Product # PA5-27195) diluted at 1:500.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
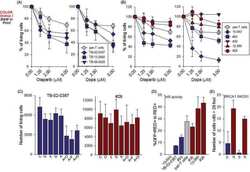
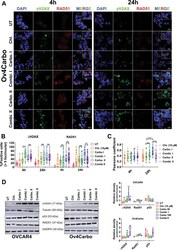
- Fig. 6 Subcellular analysis of gammaH2AX, RAD51 and p53 as markers of DNA damage/repair in Ov4Carbo (resistant) ovarian cancer cells. A Representative immunofluorescent staining of gammaH2AX and RAD51, B foci quantification and C dot plot for Pearson''s correlation coefficients representing co-localisation analysis after 4 and 24 h of treatment. ( D ) Representative immunoblots and densiometric analysis (normalised to tubulin or GADPH), depicting gammaH2AX phosphorylation (Ser139), p53 and RAD51 expression. Cells were treated with chloroxine (10 muM), carboplatin I (50 muM), combi. I (chloroxine 10 muM + carboplatin 50 muM), carbo II (100 muM) or combi. II (chloroxine 10 muM + carboplatin 100 muM). Untreated cells were used as control. Quantification was expressed as percentage of cells that showed more than five gammaH2AX or RAD51 foci/nuclei at each time point. Data show mean +- s.d., of individual ROI of duplicate coverslips. At least 100 cells were quantified for each independent experiment (IF and western blot: n >= 3 biological repeats). IF: two-way ANOVA with Tukey post hoc test; **** P < 0.0001, *** P < 0.001, ** P < 0.01 and P > 0.05 (ns). WB: unpaired t test, *** P < 0.001 ** P < 0.01, * P < 0.05 (compared to untreated) and ## P < 0.01, # P < 0.05 compared to carboplatin treated at each time point.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
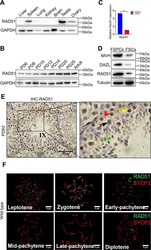
- Fig. 1 RAD51 is highly expressed in the mouse testis, predominantly in germ cells. A Western blotting was performed to detect RAD51 expression in different tissues of adult mice and GAPDH was used as the loading control. B RAD51 expression in different developmental stages of spermatogenesis was measured by Western blotting. C , D Primary isolation of spermatogenic cells and somatic cells from PD9 mouse testes. RT-qPCR ( C ) and western blotting ( D ) were conducted to test the expression of RAD51 and germ cell markers MVH and DAZL in the fraction of spermatogenic cells (FSPCs) and the fraction of somatic cells (FSCs). Data are presented as means +- S.D. ( n = 3 biologically independent samples for RT-qPCR assay). *** p < 0.001. E Immunohistochemistry staining for RAD51 was analyzed in PD62 mouse testes. (red triangle: RAD51 positive cells; L: leptotene spermatocyte; P: pachytene spermatocyte; ES: elongating spermatid). Scale bar is 50 mum. F Immunofluorescence of RAD51 (green fluorescence) and SYCP3 (red fluorescence) on chromosome spreads in wild type. Scale bar, 5 mum.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
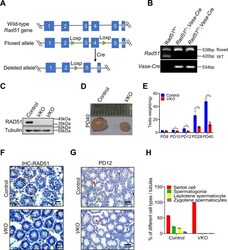
- Conditional disruption of RAD51 in germ cells results in sterility. A A schematic diagram illustrates the generation of the Rad51 conditional knockout mice. The strategy was to insert loxp sites to flank exons 3 and 4 of the mouse Rad51 gene. Cre recombinase mediated the removal of the floxed sequence to create a allele. B Genotype identification of Rad51 conditional knockout mice was analyzed by PCR of tail biopsy DNA. C The knockout efficiency was determined by western blotting in PD40 testes of RAD51-VKO mice compared to their littermate controls. D The size of testes was compared in RAD51-VKO males compared to their littermate controls from PD40 mice. E The testes weights at PD8 to PD40 were measured in RAD51-VKO mice and their littermate controls ( n = 5). Data are presented as means +- S.D. * p < 0.05, ** p < 0.01, *** p < 0.001. F Immunohistochemical staining of mouse seminiferous tubules shows the efficiency of PD8 RAD51-VKO mice and their littermate control. Scale bars are 20 mum. G Haematoxylin staining was conducted in seminiferous tubules to analyze the number and type of testicular cells in PD12 RAD51-VKO mice and their littermate controls. Scale bar, 50 mum. (red triangles: Sertoli cell; green triangles: spermatogonia; yellow triangles: leptotene spermatocyte; gray triangles: zygotene spermatocytes). H Statistics of label the different types of germ and somatic cell types to A.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
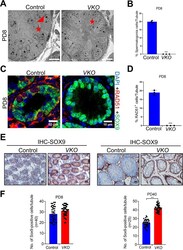
- Fig. 3 RAD51 deletion led to Sertoli cell-only syndrome in male mice. A Transmission electron microscopy (TEM) was performed to examine the detailed structure of seminiferous tubules at PD8 in VKO mice and their littermate controls (red triangles: spermatogonia; red stars: Sertoli cells). Scale bar is 10 mum. B Statistics of the germ cells to ( A ). Data are presented as means +- S.D. *** p < 0.001. C Immunofluorescence staining of the Sertoli cell marker SOX9 (green fluorescence) and RAD51 (red fluorescence) was measured in the seminiferous tubules of RAD51-VKO and control testes at PD8. Scale bar is 10 mum. D Statistics of the RAD51 positive cells to ( C ). Data are presented as means +- S.D. *** p < 0.001. E , F Immunohistochemical staining of SOX9 was analyzed in the seminiferous tubules of VKO and control mice at PD8 and PD40. SOX9 positive cells were counted using ImageJ. Scale bar is 20 mum. Data are presented as means +- S.D. * p < 0.05, ** p < 0.01, *** p < 0.001.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
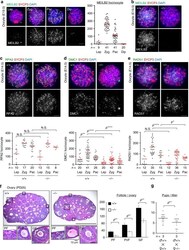
- Fig. 5 Subfertility in Meilb2 -/- female mice. a Wild-type (WT) oocytes form E19.5 mice stained with the indicated antibodies and 4,6-diamidino-2-phenylindole (DAPI). Each meiotic prophase I substage is shown. The graph shows the number of MEILB2 foci associated with the chromosome axes. The mean value is shown as a red bar. n shows the analyzed oocyte number pooled from three mice. Scale bar, 5 mum. b Zygotene oocytes from WT (+/+) and Meilb2 KO (-/-) females stained with the indicated antibodies and DAPI. Scale bar, 5 mum. c - e Zygotene oocytes from E14.5 WT (+/+) and Meilb2 KO (-/-) females stained with SYCP3 in red, DAPI in blue, and RPA2 ( c ), DMC1 ( d ), or RAD51 ( e ) in green. The images in the other stages are shown in Supplementary Fig. 7a . The graph shows the number of RPA2 ( c ), DMC1 ( d ), or RAD51 ( e ) foci associated with the chromosome axes. The mean value is shown as a red bar. n shows the analyzed oocyte number pooled from two mice for each genotype. Scale bars, 5 mum. f Ovary sections from PD25 WT (+/+) and Meilb2 KO (-/-) female mice stained with hematoxylin and eosin. The representative images of a primordial follicle (PF), primary follicle (PriF), and growing follicle (GF) are magnified. The graph shows the number of follicles in each ovary. The mean values of three independent experiments from three different mice are shown. Error bars show SD. Scale bars, 250 and 50 mum (magnified panel). g The average number of pups per lit
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
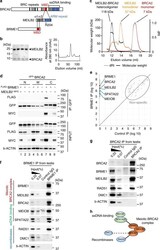
- Fig. 2 Characterization of the BRCA2-MEILB2-BRME1 ternary complex. a Schematic of interactions between BRCA2, MEILB2, and BRME1. MBD; MEILB2-binding domain, ARM repeat; armadillo-repeat domain. b Coomassie-stained gel of the heterocomplex of MEILB2 (a.a. 87-338; 27.8 kDa) and BRCA2 (a.a. 2219-2285; 10.3 kDa). The peak fractions from the size-exclusion chromatography (right graph) are shown. Purification of the individual proteins is shown in Supplementary Fig. 2a . c SEC-MALS analysis of MEILB2 (a.a. 87-338), BRCA2 (a.a. 2219-2285), and their heterocomplex. Differential refractive index (dRI) profiles are overlaid with fitted molecular weights. d IP with the GFP antibody from B16-F1 cells expressing FLAG- BRME1 and MEILB2 -MYC and GFP- BRCA2 truncations; N (a.a. 1-981), M (a.a. 982-2035), and C (a.a. 2036-3329). e Quantitative mass spectrometry of BRME1 IP (vertical axis) and control IgG IP (horizontal axis). Combined peptide intensities are plotted for each protein. Genes involved in meiotic HR regulation are indicated. Note that the other DNA repair factors, such as RAD51, were not detected by the mass spectrometry analysis, likely owing to the limited sensitivity. f , g IPs from mouse testis extracts with the BRME1 (F) or BRCA2 (G) antibodies. Asterisk: IgG heavy chain. h Protein interaction map, identified by mass spectrometry and western blotting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
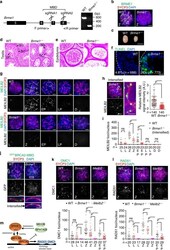
- Fig. 4 BRME1 is a stabilizer of MEILB2-BRCA2. a Schematic of the Brme1 knockout allele. Rectangle; exon. The two sgRNA-targeting sites and the location of the primers used for genotyping are shown. The gel shows the PCR results for WT and Brme1 -/- alleles. b Immunostaining of zygotene spermatocytes. Scale bar: 5 mum. c Testes from WT and Brme1 -/- males. Scale bar: 5 mm. Testis d or epididymis e sections from 8-week-old WT and Brme1 -/- males stained with hematoxylin and eosin. Spermatogonia (SG), spermatocyte (SC), and spermatid (SP). Scale bar: 100 mum. f Testis sections with stage V-VI seminiferous tubules from 8-week-old WT and Brme1 -/- males stained with TUNEL and DAPI. The percentages of TUNEL-positive seminiferous tubules (those containing more than three TUNEL-positive cells) were quantified. n shows the analyzed seminiferous tubule number pooled from two mice for each genotype. Scale bar: 15 mum. g Immunostaining of spermatocytes. Scale bar: 5 mum. h Immunostaining of Brme1 -/- zygotene spermatocyte. Graph: the quantification of MEILB2 foci intensities in zygotene spermatocytes, normalized to the average value of WT. Red bars: mean value with SD. n shows the analyzed foci number pooled from seven cells from two mice for each genotype. Scale bars: 5 mum or 1 mum in the magnified panel. i The number of MEILB2 foci associated with the chromosome axes. Red bars: mean value. n shows the analyzed spermatocyte number pooled from three mice for each genotype. j Immunostain
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
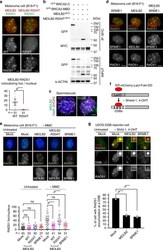
- Fig. 8 MEILB2 and BRME1 impair mitotic HR. a Mouse B16-F1 cells transfected with MEILB2 -MYC (WT) or MEILB2-R204T -MYC (R204T) stained with RAD51 and MYC antibodies. Graph: the number of MYC foci colocalizing with RAD51. n shows the analyzed RAD51-positive S to G2 phase cells. Bars: mean values with SD. b IPs with the MYC antibody from B16-F1 cells expressing MEILB2 -MYC or MEILB2-R204T -MYC with GFP- BRCA2 truncations; C (a.a. 2036-3329) or MBD (a.a. 2117-2339). c Immunostaining of zygotene spermatocytes expressing GFP- MEILB2 or GFP- MEILB2-R204T. d Mouse B16-F1 cells transfected with FLAG- BRME1 (left), MEILB2 -MYC (middle), or both FLAG- BRME1 and MEILB2 -MYC (right) and stained with RAD51 and FLAG (for BRME1) or MYC (for MEILB2) antibodies. e Mouse B16-F1 cells with or without MMC treatment transfected with MEILB2 -MYC (WT or R204T) or FLAG- BRME1 and stained with RAD51 or MYC (for MEILB2) and FLAG (for BRME1) antibodies and DAPI. Graph: the number of RAD51 foci per cell. Red bars: mean values. n shows the analyzed cell number pooled from three independent transfections. f Schematic of the U2OS-DSB reporter cell line. g U2OS-DSB reporter cell line with or without DSB induction transfected with MEILB2 -MYC or FLAG- BRME1 and stained with the RAD51 antibody. Graph: the frequency of cells with RAD51 accumulation at the DSB. The analyzed cell numbers are 78, 70, and 68 cells for mock, MEILB2, and BRME1, respectively. The mean values of two independent experiments are shown.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry