Antibody data
- Antibody Data
- Antigen structure
- References [3]
- Comments [0]
- Validations
- Western blot [3]
- Immunocytochemistry [2]
- Other assay [2]
Submit
Validation data
Reference
Comment
Report error
- Product number
- BMS1032 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- Cathepsin L Monoclonal Antibody (33-2), eBioscience™
- Antibody type
- Monoclonal
- Antigen
- Other
- Description
- Description: Cathepsin L is a lysosomal cysteine proteinase consisting of a heavy chain of about 25 kDa and a light chain of about 5 kDa derived proteolytically from the same precursor. Cathepsin L has been shown to be produced by many different cell types. Since cathepsin L is capable of degrading protein constituents of the extracellular matrix it is assumed to play a crucial role in tumor progression and metastasis and a number of other disorders where the destruction of the extracellular matrix is the major cause of disease (e.g. rheumatoid arthritis, neurodegeneration). Applications Tested: ELISA, Immunohistochemistry (Frozen), Western Blotting.
- Reactivity
- Human
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- 33-2
- Vial size
- 100 μg
- Concentration
- 1 mg/mL
- Storage
- -20°C
Submitted references Genome-scale CRISPR screens identify host factors that promote human coronavirus infection.
TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells.
Obatoclax impairs lysosomal function to block autophagy in cisplatin-sensitive and -resistant esophageal cancer cells.
Grodzki M, Bluhm AP, Schaefer M, Tagmount A, Russo M, Sobh A, Rafiee R, Vulpe CD, Karst SM, Norris MH
Genome medicine 2022 Jan 27;14(1):10
Genome medicine 2022 Jan 27;14(1):10
TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells.
Koch J, Uckeley ZM, Doldan P, Stanifer M, Boulant S, Lozach PY
The EMBO journal 2021 Aug 16;40(16):e107821
The EMBO journal 2021 Aug 16;40(16):e107821
Obatoclax impairs lysosomal function to block autophagy in cisplatin-sensitive and -resistant esophageal cancer cells.
Yu L, Wu WK, Gu C, Zhong D, Zhao X, Kong Y, Lin Q, Chan MT, Zhou Z, Liu S
Oncotarget 2016 Mar 22;7(12):14693-707
Oncotarget 2016 Mar 22;7(12):14693-707
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Knockout of Cathepsin L was achieved by CRISPR-Cas9 genome editing using LentiArray™ Lentiviral sgRNA (Product # A32042, Assay ID CRISPR1042354_LV) and LentiArray Cas9 Lentivirus (Product # A32064). Western blot analysis of Cathepsin L was performed by loading 30 µg of A549 Cas9 (Lane 1), andA549 Cathepsin L KO (Lane 2) whole cell extracts. The samples were electrophoresed using NuPAGE™ Novex™ 4-12% Bis-Tris Protein Gel (Product # NP0322BOX). Resolved proteins were then transferred onto a nitrocellulose membrane (Product # IB23001) by iBlot® 2 Dry Blotting System (Product # IB21001). The blot was probed with Anti-Cathepsin L Monoclonal Antibody (33-2), eBioscience™ (Product # BMS1032, 1 µg/mL dilution) and Goat anti-Mouse IgG (H+L) Superclonal™ Recombinant Secondary Antibody, HRP (Product # A28177, 1:5,000 dilution) using the iBright FL 1000 (Product # A32752). Chemiluminescent detection was performed using Novex® ECL Chemiluminescent Substrate Reagent Kit (Product # WP20005). Loss of signal upon CRISPR mediated knockout (KO) using the LentiArray™ CRISPR product line confirms that antibody is specific to Cathepsin L.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
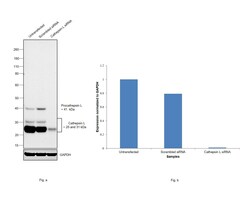
- Experimental details
- Knockdown of Cathepsin L was achieved by transfecting A549 with Cathepsin L specific siRNAs (Silencer® select Product # S223364, S3754). Western blot analysis (Fig. a) was performed using Whole cell extracts from the Cathepsin L knockdown cells (lane 3), non-targeting scrambled siRNA transfected cells (lane 2) and untransfected cells (lane 1). The blot was probed with Cathepsin L Monoclonal Antibody (33-2), eBioscience™ (Product # BMS1032, 1 µg/mL) and Goat anti-Mouse IgG (H+L) Superclonal™ Recombinant Secondary Antibody, HRP (Product # A28177, 1:4000 dilution). Densitometric analysis of all three expected bands of this western blot is shown in histogram (Fig. b). Decrease in signal upon siRNA mediated knock down confirms that antibody is specific to Cathepsin L.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
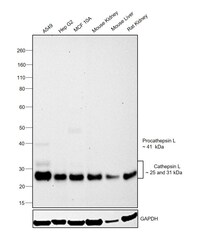
- Experimental details
- Western blot was performed using Anti-Cathepsin L Monoclonal Antibody (33-2), eBioscience™(Product # BMS1032) and a 25kDa band corresponding to the mature form of Cathepsin L was observed across cell lines and tissue extracts tested. An additional faint bands around 41kDa which corresponds to the Procathepsin L and 31kDa which also corresponds to the mature form of Cathepsin L was observed in A549. Whole cell extracts (30 µg lysate) of A549 (Lane 1), Hep G2 (Lane 2), MCF 10A (Lane 3) and tissue extracts of Mouse Kidney (Lane 4), Mouse Liver (Lane 5) and Rat Kidney (Lane 6) were electrophoresed using NuPAGE™ 4-12% Bis-Tris Protein Gel (Product # NP0322BOX). Resolved proteins were then transferred onto a Nitrocellulose membrane (Product # IB23001) by iBlot® 2 Dry Blotting System (Product # IB21001). The blot was probed with the primary antibody (1 ug/ml) and detected by chemiluminescence with Goat anti-Mouse IgG (H+L) Superclonal™ Recombinant Secondary Antibody, HRP (Product # A28177,1:4000 dilution) using the iBright FL 1000 (Product # A32752). Chemiluminescent detection was performed using Novex® ECL Chemiluminescent Substrate Reagent Kit (Product # WP20005).
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
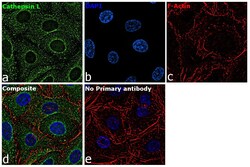
- Experimental details
- Immunofluorescence analysis of Cathepsin L was performed using 70% confluent log phase A549 cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 15 minutes, and blocked with 2% BSA for 45 minutes at room temperature. The cells were labeled with Cathepsin L Monoclonal Antibody (33-2), eBioscience™ (Product # BMS1032) at 5 µg/mL in 0.1% BSA, incubated at 4 degree celsius overnight and then labeled with Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 (Product # A32723), (1:2000), for 45 minutes at room temperature (Panel a: Green). Nuclei (Panel b:Blue) were stained with ProLong™ Diamond Antifade Mountant with DAPI (Product # P36962). F-actin (Panel c: Red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image showing cytoplasmic localization. Panel e represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
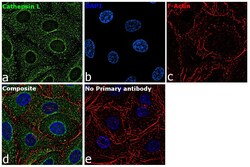
- Experimental details
- Immunofluorescence analysis of Cathepsin L was performed using 70% confluent log phase A549 cells. The cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton™ X-100 for 15 minutes, and blocked with 2% BSA for 45 minutes at room temperature. The cells were labeled with Cathepsin L Monoclonal Antibody (33-2), eBioscience™ (Product # BMS1032) at 5 µg/mL in 0.1% BSA, incubated at 4 degree celsius overnight and then labeled with Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 (Product # A32723), (1:2000), for 45 minutes at room temperature (Panel a: Green). Nuclei (Panel b:Blue) were stained with ProLong™ Diamond Antifade Mountant with DAPI (Product # P36962). F-actin (Panel c: Red) was stained with Rhodamine Phalloidin (Product # R415, 1:300). Panel d represents the merged image showing cytoplasmic localization. Panel e represents control cells with no primary antibody to assess background. The images were captured at 60X magnification.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
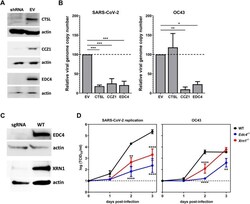
- Experimental details
- Fig. 6 Confirmation of host factor involvement by targeted shRNA knockdown and CRISPR knockout. Lentivirus-packaged shRNA clones directed to CTSL , CCZ1 , and EDC4 were transduced into HEK293T-hACE2 cells and selected with puromycin. Lentivirus-packaged sgRNA directed to EDC4 and XRN1 were transduced into SAEC-hACE2 cells and selected with puromycin. A Gene knockdown was assessed using western blotting with antibodies directed to CTSL, CCZ1, and EDC4 in cells transduced with a gene-specific shRNA or empty vector control (EV). Actin expression served as a loading control. B Triplicate wells of knockdown cells were infected with SARS-CoV-2 or OC43 at MOI 0.01. At 2 dpi, viral genome copy numbers were determined by RT-qPCR and normalized to GAPDH levels as a housekeeping control. The data are reported as the relative normalized viral genome copy number in shRNA-expressing cells compared to the EV control ( n = 3 experiments). Error bars denote standard errors of mean and P values were determined using one-way ANOVA (* P < 0.05, ** P < 0.01, *** P < 0.001). C EDC4 and XRN1 knockout in SAEC-hACE2 was assessed using western blotting with antibodies directed to EDC4 and XRN1 in cells transduced with a gene-specific sgRNA or in wild-type cells (WT). Actin expression served as a loading control. D Triplicate wells of SAEChACE2 WT, EDC4ko, and XRN1ko cells were infected with SARS-CoV-2 or OC43 and MOI 0.01. At 0hpi, 1dpi, 2dpi, and 3dpi, cell supernatants were harvested and infectious
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot ELISA
ELISA