PA5-31815
antibody from Invitrogen Antibodies
Targeting: ZNF711
CMPX1, dJ75N13.1, MRX65, MRX97, Zfp711, ZNF4, ZNF5, ZNF6
Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Other assay [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-31815 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- ZNF711 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant full-length protein
- Description
- Recommended positive controls: SK-N-SH, SK-N-SH nuclear extract, ZNF711 transfected 293T lysate. Predicted reactivity: Bovine (96%). Store product as a concentrated solution. Centrifuge briefly prior to opening the vial.
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 1 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references Characterization of the ZFX family of transcription factors that bind downstream of the start site of CpG island promoters.
Ni W, Perez AA, Schreiner S, Nicolet CM, Farnham PJ
Nucleic acids research 2020 Jun 19;48(11):5986-6000
Nucleic acids research 2020 Jun 19;48(11):5986-6000
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
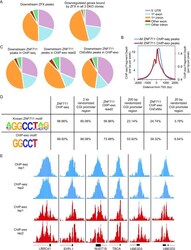
- Figure 6. Characterization of ZFX and ZNF711 binding sites. ( A ) Classification of binding sites based on genomic locations of all ZFX peaks located downstream of the TSS and ZFX downstream peaks at promoters of genes down-regulated in all 3 DKO clones. ( B ) Classification of ZNF711 downstream ChIP-seq peaks, downstream peaks identified using read2 of the ChIP-exo dataset, and downstream peaks identified by the ChexMix program (+/-10 nt from the nt identified as the binding site by the program). ( C ) Tag density plots of all ZNF711 peaks from standard ChIP-seq and from ChIP-exo. ( D ) Motif analysis using the top 5000 peaks identified from standard ZNF711 ChIP-seq (average width 1800 nt), ChIP-exo read2 (average width 300 nt), ChIP-exo by the ChexMix program (20 nt), and randomized CpG island promoter regions (width 2 kb, 200 bp, and 20 nt). The peaks were searched for the known ZNF711 motif and the 5 nt motif identified by ChIP-exo. ( E) Zoom-in comparison of peaks from ZNF711 standard ChIP-seq replicates and ChIP-exo replicates.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Other assay
Other assay