PA5-35886
antibody from Invitrogen Antibodies
Targeting: FTSJ1
CDLIV, JM23, MRX44, MRX9, SPB1, TRM7, TRMT7
Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Other assay [2]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-35886 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- FTSJ1 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant full-length protein
- Description
- Recommended positive controls: U87-MG, SK-N-SH, IMR32, SK-N-AS. Predicted reactivity: Mouse (87%), Pig (90%), Rhesus Monkey (97%), Bovine (84%). Store product as a concentrated solution. Centrifuge briefly prior to opening the vial.
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 0.78 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references Nucleoporin Nup155 is part of the p53 network in liver cancer.
Holzer K, Ori A, Cooke A, Dauch D, Drucker E, Riemenschneider P, Andres-Pons A, DiGuilio AL, Mackmull MT, Baßler J, Roessler S, Breuhahn K, Zender L, Glavy JS, Dombrowski F, Hurt E, Schirmacher P, Beck M, Singer S
Nature communications 2019 May 14;10(1):2147
Nature communications 2019 May 14;10(1):2147
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
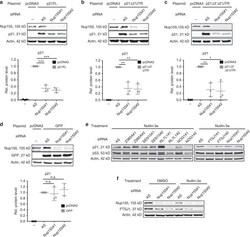
- Experimental details
- Fig. 3 Nup155-dependent p21 mRNA translation is independent of the 3'/5'UTR and potentially involves FTSJ1. a - d H1299 cells were treated either with control (AS) or two different Nup155 siRNAs (Nup155#1 and Nup155#2) for 72 h and either co-transfected with a control vector (pcDNA3) or different p21 expression constructs with p21 full-length (p21FL) ( a ) or constructs that lack either the 3'UTR (p21Delta3'UTR) ( b ) or the 5'UTR and the 3'UTR (p21Delta3'Delta5'UTR) ( c ). A GFP-expressing construct served as a negative control ( d ). Cell extracts were analysed by immunoblotting with the indicated antibodies (upper panels) and corresponding densitometric analyses derived from three independent experiments are shown in the lower panels and are normalised to the control siRNA condition. e Candidates with a suggested role in mRNA translation and downregulated upon Nup155 knockdown as revealed by the proteomic approach (see Fig. 1A ) were included in a focused RNA i approach. Reduced p21 protein accumulation and unaltered p53 levels as assayed by immunoblotting were considered as phenocopy of Nup155 depletion. Corresponding immunoblots show p21 and p53 levels (under Nutlin-3a treatment) either from the control siRNA (AS) condition or the candidate knockdown conditions (using two different siRNAs #1 and #2). Actin served as loading control. f HepG2 cells were treated either with control siRNA (AS) or two Nup155 siRNAs (Nup155#1 and Nup155#2) for 72 h and p53 was induced by addin
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
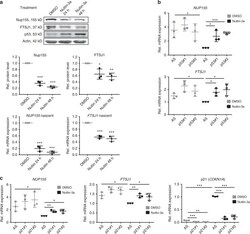
- Experimental details
- Fig. 6 Nup155 and FTSJ1 are targets of p53-mediated repression. a Sk-Hep1 cells were treated either with DMSO or Nutlin-3a for 24 h and 48 h. Cell extracts were analysed by immunoblotting with indicated antibodies (upper panel). Nup155 and FTSJ1 densitometric analyses of immunoblots derived from three independent experiments (middle panels) normalised to the DMSO condition. Relative nascent mRNA levels of NUP155 and FTSJ1 as measured by qRT-PCR (lower panels). Data are derived from three independent experiments and normalised to the DMSO condition. b Sk-Hep1 cells were treated either with control siRNA (AS) or two different p53 siRNAs (p53#1 and p53#2) for 72 h. Cells were harvested upon 48 h of Nutlin-3a treatment and extracts were analysed by qRT-PCR ( NUP155 upper panel; FTSJ1 lower panel). Data are derived from three independent experiments and normalised to the Nutlin-3a control siRNA condition. c Sk-Hep1 cells were treated either with control siRNA (AS) or two different p21 siRNAs (p21#1 and p21#2) for 72 h. Cells were harvested upon 24 h of Nutlin-3a treatment and extracts were analysed by qRT-PCR ( NUP155 left panel; FTSJ1 middle panel; p21 ( CDKN1A ) lower panel). Data are derived from three independent experiments and normalised to the Nutlin-3a control siRNA condition. * p < 0.05, ** p < 0.01, *** p < 0.001 (Student's t -test); Data are presented as mean +- stdv. Source data are provided as a Source Data file
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Other assay
Other assay