Antibody data
- Antibody Data
- Antigen structure
- References [7]
- Comments [0]
- Validations
- Immunocytochemistry [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA001648 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA001648, RRID:AB_1078635
- Product name
- Anti-DDX3X
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human DDX3X, Gene description: DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked, Alternative Gene Names: DBX, DDX14, DDX3, HLP2, Validated applications: WB, IHC, ICC, Uniprot ID: O00571, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human, Mouse, Rat
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.1 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references Aberrant cortical development is driven by impaired cell cycle and translational control in a DDX3X syndrome model
C9orf72 arginine-rich dipeptide repeat proteins disrupt karyopherin-mediated nuclear import
Pathogenic DDX3X Mutations Impair RNA Metabolism and Neurogenesis during Fetal Cortical Development
Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS
Systematic Interrogation of 3q26 Identifies TLOC1 and SKIL as Cancer Drivers
Systematic validation of antibody binding and protein subcellular localization using siRNA and confocal microscopy
Tissue profiling of the mammalian central nervous system using human antibody-based proteomics.
Hoye M, Calviello L, Poff A, Ejimogu N, Newman C, Montgomery M, Ou J, Floor S, Silver D
eLife 2022;11
eLife 2022;11
C9orf72 arginine-rich dipeptide repeat proteins disrupt karyopherin-mediated nuclear import
Hayes L, Duan L, Bowen K, Kalab P, Rothstein J
eLife 2020;9
eLife 2020;9
Pathogenic DDX3X Mutations Impair RNA Metabolism and Neurogenesis during Fetal Cortical Development
Lennox A, Hoye M, Jiang R, Johnson-Kerner B, Suit L, Venkataramanan S, Sheehan C, Alsina F, Fregeau B, Aldinger K, Moey C, Lobach I, Afenjar A, Babovic-Vuksanovic D, Bézieau S, Blackburn P, Bunt J, Burglen L, Campeau P, Charles P, Chung B, Cogné B, Curry C, D’Agostino M, Di Donato N, Faivre L, Héron D, Innes A, Isidor B, Keren B, Kimball A, Klee E, Kuentz P, Küry S, Martin-Coignard D, Mirzaa G, Mignot C, Miyake N, Matsumoto N, Fujita A, Nava C, Nizon M, Rodriguez D, Blok L, Thauvin-Robinet C, Thevenon J, Vincent M, Ziegler A, Dobyns W, Richards L, Barkovich A, Floor S, Silver D, Sherr E
Neuron 2020;106(3):404-420.e8
Neuron 2020;106(3):404-420.e8
Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS
Kamelgarn M, Chen J, Kuang L, Arenas A, Zhai J, Zhu H, Gal J
Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2016;1862(10):2004-2014
Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2016;1862(10):2004-2014
Systematic Interrogation of 3q26 Identifies TLOC1 and SKIL as Cancer Drivers
Hagerstrand D, Tong A, Schumacher S, Ilic N, Shen R, Cheung H, Vazquez F, Shrestha Y, Kim S, Giacomelli A, Rosenbluh J, Schinzel A, Spardy N, Barbie D, Mermel C, Weir B, Garraway L, Tamayo P, Mesirov J, Beroukhim R, Hahn W
Cancer Discovery 2013;3(9):1044-1057
Cancer Discovery 2013;3(9):1044-1057
Systematic validation of antibody binding and protein subcellular localization using siRNA and confocal microscopy
Stadler C, Hjelmare M, Neumann B, Jonasson K, Pepperkok R, Uhlén M, Lundberg E
Journal of Proteomics 2012;75(7):2236-2251
Journal of Proteomics 2012;75(7):2236-2251
Tissue profiling of the mammalian central nervous system using human antibody-based proteomics.
Mulder J, Björling E, Jonasson K, Wernérus H, Hober S, Hökfelt T, Uhlén M
Molecular & cellular proteomics : MCP 2009 Jul;8(7):1612-22
Molecular & cellular proteomics : MCP 2009 Jul;8(7):1612-22
No comments: Submit comment
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
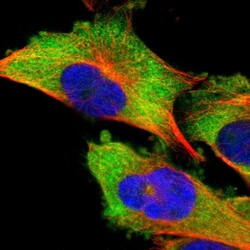
- Experimental details
- Immunofluorescent staining of human cell line U-251 MG shows localization to cytosol.
- Sample type
- Human
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry