PA5-100756
antibody from Invitrogen Antibodies
Targeting: HIVEP2
HIV-EP2, MBP-2, MIBP1, Schnurri-2, ZAS2, ZNF40B
Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Other assay [2]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-100756 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- HIVEP2 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Synthetic peptide
- Description
- Antibody detects endogenous levels of total MIBP1.
- Reactivity
- Human, Mouse, Rat
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 1 mg/mL
- Storage
- -20°C
Submitted references EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma.
Fang R, Chen X, Zhang S, Shi H, Ye Y, Shi H, Zou Z, Li P, Guo Q, Ma L, He C, Huang S
Nature communications 2021 Jan 8;12(1):177
Nature communications 2021 Jan 8;12(1):177
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
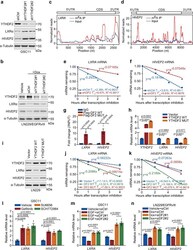
- Fig. 6 YTHDF2 downregulates LXRA and HIVEP2 through m 6 A-dependent mRNA decay. a Western blotting of LXRalpha and HIVEP2 in GSC11 cells stably transfected with control shRNA or two YTHDF2 shRNAs. b Western blotting of LXRalpha and HIVEP2 in LN229/EGFRvIII tet-on cells cultured with vehicle or Doxycycline and transfected with control siRNA or two individual YTHDF2 siRNAs. c , d MeRIP-seq of GSC11 cells shows m 6 A peaks at individual mRNAs of LXRA and HIVEP2 . The y -axis shows normalized reads distribution for input (blue) and m 6 A IP (red) along the transcripts. The schematic representation of mRNA of LXRA or HIVEP2 (top panel). e , f Lifetime of LXRA ( e ) and HIVEP2 ( f ) mRNA in GSC11 cells expressing shYTHDF2 (shDF2), or shControl (shCtrl). Transcription was inhibited by actinomycin D. g CLIP-qPCR showing the association of LXRA and HIVEP2 transcripts with wild-type (WT) or m 6 A recognition defective (MUT) Flag-YTHDF2 in LN229 cells. Data are mean +- S.E.M., n = 4 biologically independent experiments. h mRNA levels of YTHDF2 , LXRA , and HIVEP2 in LN229 cells transfected with vector, wild type (WT) or m 6 A recognition defective (MUT) YTHDF2. Data are mean +- S.E.M., n = 3 biologically independent experiments. i Western blotting of YTHDF2, LXRalpha, and HIVEP2 in LN229 cells transfected with vector, wild-type (WT) or recognition defective (MUT) YTHDF2. j , k Lifetime of LXRA ( j ) or HIVEP2 ( k ) mRNA in LN229 cells expressing wild-type (WT) or m 6 A recognition defec
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
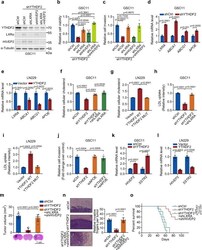
- Fig. 7 LXRalpha and HIVEP2 are functionally essential targets of YTHDF2 in cell proliferation, invasion, cholesterol dysregulation, and tumorigenesis of GBM cells. a Western blotting of YTHDF2, LXRalpha, and HIVEP2 in GSC11 cells stably expressing shCtrl, shYTHDF2, or shYTHDF2 plus shLXRA or/and shHIVEP2. Representative blot of three independent experiments is shown. b Proliferation of the cells in ( a ) was measured. Data are mean +- S.E.M., n = 6 biologically independent experiments. c In vitro invasion assay for GSC11 cells expressing shCtrl, shYTHDF2, or shYTHDF2 plus shLXRA or/and shHIVEP2. Data are mean +- S.E.M., n = 4 wells examined over three independent experiments. d , e mRNA levels of LXRA and its downstream targets in shCtrl and shYTHDF2 GSC11 cells ( d ) or in LN229 cells expressing vector or YTHDF2 plasmid ( e ). Data are mean +- S.E.M., n = 3 biologically independent experiments (unpaired two-sided t test). f , g Relative cellular cholesterol of GSC11 cells stably expressing shCtrl, shYTHDF2, shLXRA, or shYTHDF2 plus shLXRA ( f ) or LN229 cells expressing vector or YTHDF2 plasmid ( g ). Data are mean +- S.E.M., n = 6 ( f ) or n = 3 ( g ) biologically independent experiments. h , i Quantification of LDL uptake in GSC11 cells expressing shCtrl, shYTHDF2, or shYTHDF2 plus shLXRA ( h ) or in LN229 cells expressing wild-type (WT) or m 6 A recognition defective (MUT) YTHDF2 ( i ). Data are mean +- S.E.M., n = 3 biologically independent experiments. j In vitro invasi
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Other assay
Other assay