Antibody data
- Antibody Data
- Antigen structure
- References [4]
- Comments [0]
- Validations
- Immunocytochemistry [1]
- Other assay [6]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-66827 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- SRRM2 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant protein fragment
- Description
- Immunogen sequence: LMLLEKDVNPG GKEETPGQRP AVTETHQLAE LNEKKNERLR AAFGISDSYV DGSSFDPQRR AREAKQPAPE P Highest antigen sequence identity to the following orthologs - mouse 96%, rat 97%.
- Reactivity
- Human
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 0.10 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references RNA is required for the integrity of multiple nuclear and cytoplasmic membrane-less RNP granules.
Revealing RCOR2 as a regulatory component of nuclear speckles.
Identification of abscission checkpoint bodies as structures that regulate ESCRT factors to control abscission timing.
SON and SRRM2 are essential for nuclear speckle formation.
Decker CJ, Burke JM, Mulvaney PK, Parker R
The EMBO journal 2022 May 2;41(9):e110137
The EMBO journal 2022 May 2;41(9):e110137
Revealing RCOR2 as a regulatory component of nuclear speckles.
Rivera C, Verbel-Vergara D, Arancibia D, Lappala A, González M, Guzmán F, Merello G, Lee JT, Andrés ME
Epigenetics & chromatin 2021 Nov 24;14(1):51
Epigenetics & chromatin 2021 Nov 24;14(1):51
Identification of abscission checkpoint bodies as structures that regulate ESCRT factors to control abscission timing.
Strohacker LK, Mackay DR, Whitney MA, Couldwell GC, Sundquist WI, Ullman KS
eLife 2021 Aug 4;10
eLife 2021 Aug 4;10
SON and SRRM2 are essential for nuclear speckle formation.
Ilik İA, Malszycki M, Lübke AK, Schade C, Meierhofer D, Aktaş T
eLife 2020 Oct 23;9
eLife 2020 Oct 23;9
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
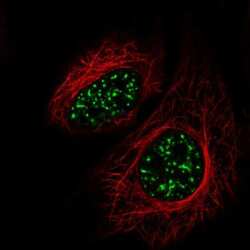
- Experimental details
- Immunofluorescent staining of SRRM2 in human cell line HeLa shows localization to nuclear speckles. Samples were probed using a SRRM2 Polyclonal Antibody (Product # PA5-66827).
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
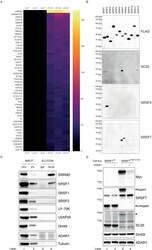
- Experimental details
- Figure 1. SC35 mAb immunoprecipitation followed by MS identifies SRRM2 as the top hit. ( A ) The Top50 hits identified by the MS are depicted on a heatmap showing the number of unique peptides detected for each protein. Also see Figure 1--figure supplement 1A for an intensity vs MS/MS spectra plot and Supplementary file 2 for raw MaxQuant results ( B ) Streptavidin pull-down of biotin tagged ectopically expressed SRSF proteins 1 to 12 are blotted with FLAG antibody in order to show the amounts of loaded proteins on PAGE. Western blot using mAb SC35 detects SRSF7 with highest sensitivity in comparison to all other SRSF proteins, but also weakly reacts with SRSF1, 2, and 12. Specific antibodies against SRSF2 and SRSF7 are used to validate the authenticity of the purified proteins from stable cell lines in corresponding lanes and blots. ( C ) SC35-IP performed on lysates from wild-type HEK293 cells identifies SRRM2 as the most enriched protein with a weaker enrichment for SRSF7 but no enrichment for SRSF1 or SRSF2 using western blotting (compare Lane 4 across blots). ( D ) Homozygous knock-in of the 2xMyc-FKBP12 F36V tag into SRSF7 gene locus shifts the SC35 band from 35 kDa to 50 kDa (compare Lanes 1 and 3 on the SC35 blot) and upon induction of degradation with dTAG7 the shifted band is lost (compare Lanes 3 and 4 in Myc, SRSF7, and SC35 blots). This blot validates that the 35 kDa band identified by mAb SC35 blots corresponds to SRSF7. Figure 1--figure supplement 1. SC35 pull-
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
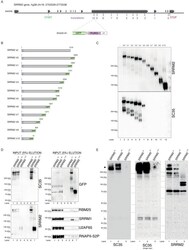
- Experimental details
- Figure 2. Endogenous truncating mutations of SRRM2 prove mAb SC35 as an SRRM2 antibody. ( A ) The strategy for the CRISPaint generated endogenous truncating mutations (0-to-10) accompanied by the TagGFP2 (depicted as GFP for simplicity) fusion are shown. ( B ) The sizes of SRRM2 truncated GFP fusion proteins are displayed. ( C ) Protein purified using a GFP-trap pull-down from lysates of corresponding stable HAP1 cell lines carrying the truncated SRRM2 alleles are run on PAGE. Western blotting of SRRM2 using an antibody generated against the common N-terminus is used to show the amount of loaded protein on the gel. SC35 blot shows a significant reduction in signal intensity of SRRM2-tr5 and a complete loss of signal from SRRM2-tr6 to tr10. ( D ) GFP-trap pull-down performed on lysates from wild-type, tr0 and tr10 HAP1 cells enrich for SRRM2 in tr0 cells, indicating the GFP-tagged allele is specific to SRRM2 and is detected by also SC35 blot (Lanes 1 and 2 inputs compared to Lanes 5 and 6 on the upper and lower left-side blots). SRRM2 also co-purifies two other NS-associated proteins; SRRM1 and RBM25 (Lane 6 on lower right-side blots). SRRM2-tr10 is not detected by SC35 but the pull-down efficiency (Lane 7 on upper left-side blot) and loading is validated by SRRM2 (Lanes 3 and 7 on lower left-side blot) and GFP blots (Lanes 3 and 7 on upper left-side blot). ( E ) Total cell lysates from wild-type, tr0 and tr10 HAP1 cells are run on 4-12% polyacrylamide gel and blotted with SC3
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- 3 Figure The structure of nucleoli, nuclear speckles and Cajal bodies is dependent on nuclear RNA Analysis of nuclear RNA granules in A549 cells with RNase L targeted to the nucleus (NLS-RL-WT) either mock transfected or treated with poly(I:C) (PIC) for 4 to 5 h. FISH analysis to detect poly(A)+ RNA and nucleolar-localized snoRD3A RNA. IF analysis to detect nucleolar granular component protein nucleophosmin (NPM1). Graph of the mean and value of the average volume of individual snoRD3A foci in nuclei. 17 nuclei mock-treated cells, 31 nuclei PIC-treated cells. Fraction of nuclei with NPM1 protein enriched in ring structures classified as granular component assemblies or dispersed in nucleoplasm. 17 nuclei mock-treated cells, 31 nuclei PIC-treated cells. IF analysis to detect dense fibrillar component protein, ribosome processing factor 1 (RPF1). Graph of the mean and value of the average volume of individual snoRD3A foci in nuclei. 43 nuclei mock-treated cells, 69 nuclei PIC-treated cells. Graph of the mean and value of the average volume of RPF1 foci in 43 mock-treated cells and 69 nuclei PIC-treated cells. FISH analysis to detect poly(A)+ RNA and nuclear speckle-localized MALAT1 RNA and IF analysis with anti-sc35 antibody to detect nuclear speckle protein SRRM2. Graph of the mean and value of the intensity of MALAT1 signal divided by the nuclear volume of individual nuclei. 51 nuclei mock-treated cells, 56 nuclei PIC-treated cells. Graph of the median and value of the volume
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
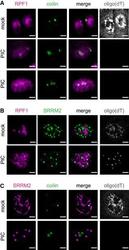
- 4 Figure Protein components of nuclear RNA granules do not co-assemble when nuclear RNA is degraded Analysis of nuclear RNA granules in A549 cells with RNase L targeted to the nucleus (NLS-RL-WT) either mock transfected or treated with poly(I:C) (PIC) for 5 h. Oligo(dT) FISH and IF analysis of nucleolar protein RPF1 and Cajal body protein coilin. Oligo(dT) FISH and IF analysis of nucleolar protein RPF1 and nuclear speckle protein SRRM2. Oligo(dT) FISH and IF analysis of Cajal body protein coilin and nuclear speckle protein SRRM2. Data information: Scale bar 5 microns.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
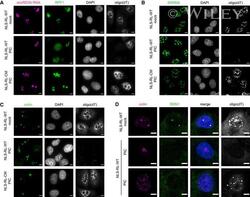
- EV4 Figure Changes in nuclear RNA granule morphology is dependent on loss of nuclear RNA and SMN1 disperses into nucleoplasm in response to nuclear RNA degradation IF analysis of nuclear RNA granule proteins in A549 cells expressing nuclear-localized wild-type RNase L or catalytic mutant RNase L-R667A mock transfected or treated with poly(I:C) for 5 h. FISH analysis to detect nucleolar-localized snoRD3A RNA and poly(A)+ RNA. IF analysis of nucleolar protein RPF1. Oligo(dT) FISH and IF analysis of nuclear speckle protein SRRM2 with sc35 antibody. Oligo(dT) FISH and IF analysis of Cajal body protein coilin. Oligo(dT) FISH and IF analysis of Cajal body protein coilin and SMN1. Data information: Scale bar 5 microns.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
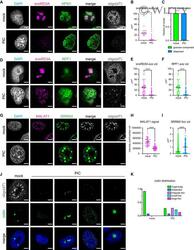
- Experimental details
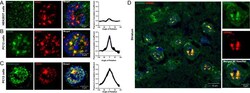
- Fig. 2 RCOR2 is recruited to nuclear speckles in the mouse brain. A , B Comparison of colocalization between RCOR2 (green) and SRRM2 (red) in HEK293T ( A ) and PC12 ( B ) cells. Right-side plots show Van Steensel's colocalization test profiles. C Colocalization analysis between overexpressed FLAG-tagged RCOR2 (green) and SRRM2 (red) in PC12 cells. Right-side plot shows Van Steensel's colocalization test. D Tissue immunofluorescence of SRRM2 (red) and RCOR2 (green) in mice striatum slices. The right panels show a zoomed-in nucleus, illustrating speckles found in single nuclei of the brain tissues
 Explore
Explore Validate
Validate Learn
Learn Immunocytochemistry
Immunocytochemistry