Antibody data
- Antibody Data
- Antigen structure
- References [15]
- Comments [0]
- Validations
- Western blot [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 23111-1-AP - Provider product page

- Provider
- Proteintech Group
- Proper citation
- Proteintech Cat#23111-1-AP, RRID:AB_2744527
- Product name
- ZRANB3 antibody
- Antibody type
- Polyclonal
- Description
- KD/KO validated ZRANB3 antibody (Cat. #23111-1-AP) is a rabbit polyclonal antibody that shows reactivity with human and has been validated for the following applications: IF, IHC, WB, ELISA.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 20ul, 150ul
Submitted references RAD52 prevents accumulation of Polα -dependent replication gaps at perturbed replication forks in human cells.
RFWD3 promotes ZRANB3 recruitment to regulate the remodeling of stalled replication forks.
Excessive reactive oxygen species induce transcription-dependent replication stress.
DDX17 helicase promotes resolution of R-loop-mediated transcription-replication conflicts in human cells.
Active DNA damage eviction by HLTF stimulates nucleotide excision repair.
p53 isoforms differentially impact on the POLι dependent DNA damage tolerance pathway.
BRCA2 associates with MCM10 to suppress PRIMPOL-mediated repriming and single-stranded gap formation after DNA damage.
Sequential role of RAD51 paralog complexes in replication fork remodeling and restart.
Fork Cleavage-Religation Cycle and Active Transcription Mediate Replication Restart after Fork Stalling at Co-transcriptional R-Loops.
WRNIP1 Protects Reversed DNA Replication Forks from SLX4-Dependent Nucleolytic Cleavage.
Rad52 prevents excessive replication fork reversal and protects from nascent strand degradation.
BET Inhibition Induces HEXIM1- and RAD51-Dependent Conflicts between Transcription and Replication.
CtIP-Mediated Fork Protection Synergizes with BRCA1 to Suppress Genomic Instability upon DNA Replication Stress.
Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity.
Replication fork reversal triggers fork degradation in BRCA2-defective cells.
Di Biagi L, Marozzi G, Malacaria E, Honda M, Aiello FA, Valenzisi P, Spies M, Franchitto A, Pichierri P
bioRxiv : the preprint server for biology 2024 Aug 17;
bioRxiv : the preprint server for biology 2024 Aug 17;
RFWD3 promotes ZRANB3 recruitment to regulate the remodeling of stalled replication forks.
Moore CE, Yalcindag SE, Czeladko H, Ravindranathan R, Wijesekara Hanthi Y, Levy JC, Sannino V, Schindler D, Ciccia A, Costanzo V, Elia AEH
The Journal of cell biology 2023 May 1;222(5)
The Journal of cell biology 2023 May 1;222(5)
Excessive reactive oxygen species induce transcription-dependent replication stress.
Andrs M, Stoy H, Boleslavska B, Chappidi N, Kanagaraj R, Nascakova Z, Menon S, Rao S, Oravetzova A, Dobrovolna J, Surendranath K, Lopes M, Janscak P
Nature communications 2023 Mar 30;14(1):1791
Nature communications 2023 Mar 30;14(1):1791
DDX17 helicase promotes resolution of R-loop-mediated transcription-replication conflicts in human cells.
Boleslavska B, Oravetzova A, Shukla K, Nascakova Z, Ibini ON, Hasanova Z, Andrs M, Kanagaraj R, Dobrovolna J, Janscak P
Nucleic acids research 2022 Nov 28;50(21):12274-12290
Nucleic acids research 2022 Nov 28;50(21):12274-12290
Active DNA damage eviction by HLTF stimulates nucleotide excision repair.
van Toorn M, Turkyilmaz Y, Han S, Zhou D, Kim HS, Salas-Armenteros I, Kim M, Akita M, Wienholz F, Raams A, Ryu E, Kang S, Theil AF, Bezstarosti K, Tresini M, Giglia-Mari G, Demmers JA, Schärer OD, Choi JH, Vermeulen W, Marteijn JA
Molecular cell 2022 Apr 7;82(7):1343-1358.e8
Molecular cell 2022 Apr 7;82(7):1343-1358.e8
p53 isoforms differentially impact on the POLι dependent DNA damage tolerance pathway.
Guo Y, Rall-Scharpf M, Bourdon JC, Wiesmüller L, Biber S
Cell death & disease 2021 Oct 13;12(10):941
Cell death & disease 2021 Oct 13;12(10):941
BRCA2 associates with MCM10 to suppress PRIMPOL-mediated repriming and single-stranded gap formation after DNA damage.
Kang Z, Fu P, Alcivar AL, Fu H, Redon C, Foo TK, Zuo Y, Ye C, Baxley R, Madireddy A, Buisson R, Bielinsky AK, Zou L, Shen Z, Aladjem MI, Xia B
Nature communications 2021 Oct 13;12(1):5966
Nature communications 2021 Oct 13;12(1):5966
Sequential role of RAD51 paralog complexes in replication fork remodeling and restart.
Berti M, Teloni F, Mijic S, Ursich S, Fuchs J, Palumbieri MD, Krietsch J, Schmid JA, Garcin EB, Gon S, Modesti M, Altmeyer M, Lopes M
Nature communications 2020 Jul 15;11(1):3531
Nature communications 2020 Jul 15;11(1):3531
Fork Cleavage-Religation Cycle and Active Transcription Mediate Replication Restart after Fork Stalling at Co-transcriptional R-Loops.
Chappidi N, Nascakova Z, Boleslavska B, Zellweger R, Isik E, Andrs M, Menon S, Dobrovolna J, Balbo Pogliano C, Matos J, Porro A, Lopes M, Janscak P
Molecular cell 2020 Feb 6;77(3):528-541.e8
Molecular cell 2020 Feb 6;77(3):528-541.e8
WRNIP1 Protects Reversed DNA Replication Forks from SLX4-Dependent Nucleolytic Cleavage.
Porebski B, Wild S, Kummer S, Scaglione S, Gaillard PL, Gari K
iScience 2019 Nov 22;21:31-41
iScience 2019 Nov 22;21:31-41
Rad52 prevents excessive replication fork reversal and protects from nascent strand degradation.
Malacaria E, Pugliese GM, Honda M, Marabitti V, Aiello FA, Spies M, Franchitto A, Pichierri P
Nature communications 2019 Mar 29;10(1):1412
Nature communications 2019 Mar 29;10(1):1412
BET Inhibition Induces HEXIM1- and RAD51-Dependent Conflicts between Transcription and Replication.
Bowry A, Piberger AL, Rojas P, Saponaro M, Petermann E
Cell reports 2018 Nov 20;25(8):2061-2069.e4
Cell reports 2018 Nov 20;25(8):2061-2069.e4
CtIP-Mediated Fork Protection Synergizes with BRCA1 to Suppress Genomic Instability upon DNA Replication Stress.
Przetocka S, Porro A, Bolck HA, Walker C, Lezaja A, Trenner A, von Aesch C, Himmels SF, D'Andrea AD, Ceccaldi R, Altmeyer M, Sartori AA
Molecular cell 2018 Nov 1;72(3):568-582.e6
Molecular cell 2018 Nov 1;72(3):568-582.e6
Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity.
Vujanovic M, Krietsch J, Raso MC, Terraneo N, Zellweger R, Schmid JA, Taglialatela A, Huang JW, Holland CL, Zwicky K, Herrador R, Jacobs H, Cortez D, Ciccia A, Penengo L, Lopes M
Molecular cell 2017 Sep 7;67(5):882-890.e5
Molecular cell 2017 Sep 7;67(5):882-890.e5
Replication fork reversal triggers fork degradation in BRCA2-defective cells.
Mijic S, Zellweger R, Chappidi N, Berti M, Jacobs K, Mutreja K, Ursich S, Ray Chaudhuri A, Nussenzweig A, Janscak P, Lopes M
Nature communications 2017 Oct 16;8(1):859
Nature communications 2017 Oct 16;8(1):859
No comments: Submit comment
Supportive validation
- Submitted by
- Proteintech Group (provider)
- Main image
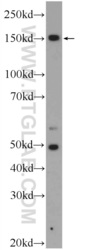
- Experimental details
- The ZRANB3 antibody from Proteintech is a rabbit polyclonal antibody to a fusion protein of human ZRANB3. This antibody recognizes human antigen. The ZRANB3 antibody has been validated for the following applications: ELISA, WB analysis.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot ELISA
ELISA