Antibody data
- Antibody Data
- Antigen structure
- References [24]
- Comments [0]
- Validations
- Immunohistochemistry [1]
- Other assay [29]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 39-2500 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- AID Monoclonal Antibody (ZA001)
- Antibody type
- Monoclonal
- Antigen
- Synthetic peptide
- Reactivity
- Human, Mouse
- Host
- Mouse
- Isotype
- IgG
- Antibody clone number
- ZA001
- Vial size
- 100 μg
- Concentration
- 0.5 mg/mL
- Storage
- -20°C
Submitted references Optimized high-fidelity 3DPCR to assess potential mitochondrial targeting by activation-induced cytidine deaminase.
Plasma cells, plasmablasts, and AID(+)/CD30(+) B lymphoblasts inside and outside germinal centres: details of the basal light zone and the outer zone in human palatine tonsils.
Disease-associated CTNNBL1 mutation impairs somatic hypermutation by decreasing nuclear AID.
The RNA-binding protein ROD1/PTBP3 cotranscriptionally defines AID-loading sites to mediate antibody class switch in mammalian genomes.
Regulation of the DNA Repair Complex during Somatic Hypermutation and Class-Switch Recombination.
The expansion in lymphoid organs of IL-4(+) BATF(+) T follicular helper cells is linked to IgG4 class switching in vivo.
Aberrant expression of AID and AID activators of NF-κB and PAX5 is irrelevant to EBV-associated gastric cancers, but is associated with carcinogenesis in certain EBV-non-associated gastric cancers.
Phosphatidylinositol 3-kinase δ blockade increases genomic instability in B cells.
Isotype-switched follicular lymphoma displays dissociation between activation-induced cytidine deaminase expression and somatic hypermutation.
Bcl6 Is Required for Somatic Hypermutation and Gene Conversion in Chicken DT40 Cells.
Aberrant activation-induced cytidine deaminase expression in Philadelphia chromosome-positive B-cell acute lymphoblastic leukemia.
Leukemic non-nodal mantle cell lymphomas have a distinct phenotype and are associated with deletion of PARP1 and 13q14.
AID expression in B-cell lymphomas causes accumulation of genomic uracil and a distinct AID mutational signature.
BCL2 mutations are associated with increased risk of transformation and shortened survival in follicular lymphoma.
B cell Rab7 mediates induction of activation-induced cytidine deaminase expression and class-switching in T-dependent and T-independent antibody responses.
Activation-induced cytidine deaminase (AID) is localized to subnuclear domains enriched in splicing factors.
The role of activation-induced cytidine deaminase expression in gastric adenocarcinoma.
Scaffold functions of 14-3-3 adaptors in B cell immunoglobulin class switch DNA recombination.
Combinatorial H3K9acS10ph histone modification in IgH locus S regions targets 14-3-3 adaptors and AID to specify antibody class-switch DNA recombination.
Activation-induced cytidine deaminase (AID)-associated multigene signature to assess impact of AID in etiology of diseases with inflammatory component.
Aberrant expression and mutation-inducing activity of AID in human lung cancer.
14-3-3 adaptor proteins recruit AID to 5'-AGCT-3'-rich switch regions for class switch recombination.
E2A-positive gastric MALT lymphoma has weaker plasmacytoid infiltrates and stronger expression of the memory B-cell-associated miR-223: possible correlation with stage and treatment response.
Increased targeting of donor switch region and IgE in Sgamma1-deficient B cells.
Wu H, Zhang K, Chen Y, Li J, Strout MP, Gu X
FEBS open bio 2020 Sep;10(9):1782-1792
FEBS open bio 2020 Sep;10(9):1782-1792
Plasma cells, plasmablasts, and AID(+)/CD30(+) B lymphoblasts inside and outside germinal centres: details of the basal light zone and the outer zone in human palatine tonsils.
Steiniger BS, Raimer L, Ecke A, Stuck BA, Cetin Y
Histochemistry and cell biology 2020 Jul;154(1):55-75
Histochemistry and cell biology 2020 Jul;154(1):55-75
Disease-associated CTNNBL1 mutation impairs somatic hypermutation by decreasing nuclear AID.
Kuhny M, Forbes LR, Çakan E, Vega-Loza A, Kostiuk V, Dinesh RK, Glauzy S, Stray-Pedersen A, Pezzi AE, Hanson IC, Vargas-Hernandez A, Xu ML, Coban-Akdemir ZH, Jhangiani SN, Muzny DM, Gibbs RA, Lupski JR, Chinn IK, Schatz DG, Orange JS, Meffre E
The Journal of clinical investigation 2020 Aug 3;130(8):4411-4422
The Journal of clinical investigation 2020 Aug 3;130(8):4411-4422
The RNA-binding protein ROD1/PTBP3 cotranscriptionally defines AID-loading sites to mediate antibody class switch in mammalian genomes.
Chen J, Cai Z, Bai M, Yu X, Zhang C, Cao C, Hu X, Wang L, Su R, Wang D, Wang L, Yao Y, Ye R, Hou B, Yu Y, Yu S, Li J, Xue Y
Cell research 2018 Oct;28(10):981-995
Cell research 2018 Oct;28(10):981-995
Regulation of the DNA Repair Complex during Somatic Hypermutation and Class-Switch Recombination.
Kumar A, Priya A, Ahmed T, Grundström C, Negi N, Grundström T
Journal of immunology (Baltimore, Md. : 1950) 2018 Jun 15;200(12):4146-4156
Journal of immunology (Baltimore, Md. : 1950) 2018 Jun 15;200(12):4146-4156
The expansion in lymphoid organs of IL-4(+) BATF(+) T follicular helper cells is linked to IgG4 class switching in vivo.
Maehara T, Mattoo H, Mahajan VS, Murphy SJ, Yuen GJ, Ishiguro N, Ohta M, Moriyama M, Saeki T, Yamamoto H, Yamauchi M, Daccache J, Kiyoshima T, Nakamura S, Stone JH, Pillai S
Life science alliance 2018 Jan;1(1)
Life science alliance 2018 Jan;1(1)
Aberrant expression of AID and AID activators of NF-κB and PAX5 is irrelevant to EBV-associated gastric cancers, but is associated with carcinogenesis in certain EBV-non-associated gastric cancers.
Mohri T, Nagata K, Kuwamoto S, Matsushita M, Sugihara H, Kato M, Horie Y, Murakami I, Hayashi K
Oncology letters 2017 Jun;13(6):4133-4140
Oncology letters 2017 Jun;13(6):4133-4140
Phosphatidylinositol 3-kinase δ blockade increases genomic instability in B cells.
Compagno M, Wang Q, Pighi C, Cheong TC, Meng FL, Poggio T, Yeap LS, Karaca E, Blasco RB, Langellotto F, Ambrogio C, Voena C, Wiestner A, Kasar SN, Brown JR, Sun J, Wu CJ, Gostissa M, Alt FW, Chiarle R
Nature 2017 Feb 23;542(7642):489-493
Nature 2017 Feb 23;542(7642):489-493
Isotype-switched follicular lymphoma displays dissociation between activation-induced cytidine deaminase expression and somatic hypermutation.
Scherer F, Navarrete MA, Bertinetti-Lapatki C, Boehm J, Schmitt-Graeff A, Veelken H
Leukemia & lymphoma 2016;57(1):151-60
Leukemia & lymphoma 2016;57(1):151-60
Bcl6 Is Required for Somatic Hypermutation and Gene Conversion in Chicken DT40 Cells.
Williams AM, Maman Y, Alinikula J, Schatz DG
PloS one 2016;11(2):e0149146
PloS one 2016;11(2):e0149146
Aberrant activation-induced cytidine deaminase expression in Philadelphia chromosome-positive B-cell acute lymphoblastic leukemia.
Shi Y, Zhao X, Durkin L, Rogers HJ, Hsi ED
Human pathology 2016 Jun;52:173-8
Human pathology 2016 Jun;52:173-8
Leukemic non-nodal mantle cell lymphomas have a distinct phenotype and are associated with deletion of PARP1 and 13q14.
Gallo M, Cacheux V, Vincent L, Bret C, Tempier A, Guittard C, Macé A, Leventoux N, Costes V, Szablewski V
Virchows Archiv : an international journal of pathology 2016 Dec;469(6):697-706
Virchows Archiv : an international journal of pathology 2016 Dec;469(6):697-706
AID expression in B-cell lymphomas causes accumulation of genomic uracil and a distinct AID mutational signature.
Pettersen HS, Galashevskaya A, Doseth B, Sousa MM, Sarno A, Visnes T, Aas PA, Liabakk NB, Slupphaug G, Sætrom P, Kavli B, Krokan HE
DNA repair 2015 Jan;25:60-71
DNA repair 2015 Jan;25:60-71
BCL2 mutations are associated with increased risk of transformation and shortened survival in follicular lymphoma.
Correia C, Schneider PA, Dai H, Dogan A, Maurer MJ, Church AK, Novak AJ, Feldman AL, Wu X, Ding H, Meng XW, Cerhan JR, Slager SL, Macon WR, Habermann TM, Karp JE, Gore SD, Kay NE, Jelinek DF, Witzig TE, Nowakowski GS, Kaufmann SH
Blood 2015 Jan 22;125(4):658-67
Blood 2015 Jan 22;125(4):658-67
B cell Rab7 mediates induction of activation-induced cytidine deaminase expression and class-switching in T-dependent and T-independent antibody responses.
Pone EJ, Lam T, Lou Z, Wang R, Chen Y, Liu D, Edinger AL, Xu Z, Casali P
Journal of immunology (Baltimore, Md. : 1950) 2015 Apr 1;194(7):3065-78
Journal of immunology (Baltimore, Md. : 1950) 2015 Apr 1;194(7):3065-78
Activation-induced cytidine deaminase (AID) is localized to subnuclear domains enriched in splicing factors.
Hu Y, Ericsson I, Doseth B, Liabakk NB, Krokan HE, Kavli B
Experimental cell research 2014 Mar 10;322(1):178-92
Experimental cell research 2014 Mar 10;322(1):178-92
The role of activation-induced cytidine deaminase expression in gastric adenocarcinoma.
Batsaikhan BE, Kurita N, Iwata T, Sato H, Yoshikawa K, Takasu C, Kashihara H, Matsumoto N, Ishibashi H, Shimada M
Anticancer research 2014 Feb;34(2):995-1000
Anticancer research 2014 Feb;34(2):995-1000
Scaffold functions of 14-3-3 adaptors in B cell immunoglobulin class switch DNA recombination.
Lam T, Thomas LM, White CA, Li G, Pone EJ, Xu Z, Casali P
PloS one 2013;8(11):e80414
PloS one 2013;8(11):e80414
Combinatorial H3K9acS10ph histone modification in IgH locus S regions targets 14-3-3 adaptors and AID to specify antibody class-switch DNA recombination.
Li G, White CA, Lam T, Pone EJ, Tran DC, Hayama KL, Zan H, Xu Z, Casali P
Cell reports 2013 Nov 14;5(3):702-714
Cell reports 2013 Nov 14;5(3):702-714
Activation-induced cytidine deaminase (AID)-associated multigene signature to assess impact of AID in etiology of diseases with inflammatory component.
Mechtcheriakova D, Sobanov Y, Holtappels G, Bajna E, Svoboda M, Jaritz M, Bachert C, Jensen-Jarolim E
PloS one 2011;6(10):e25611
PloS one 2011;6(10):e25611
Aberrant expression and mutation-inducing activity of AID in human lung cancer.
Shinmura K, Igarashi H, Goto M, Tao H, Yamada H, Matsuura S, Tajima M, Matsuda T, Yamane A, Funai K, Tanahashi M, Niwa H, Ogawa H, Sugimura H
Annals of surgical oncology 2011 Jul;18(7):2084-92
Annals of surgical oncology 2011 Jul;18(7):2084-92
14-3-3 adaptor proteins recruit AID to 5'-AGCT-3'-rich switch regions for class switch recombination.
Xu Z, Fulop Z, Wu G, Pone EJ, Zhang J, Mai T, Thomas LM, Al-Qahtani A, White CA, Park SR, Steinacker P, Li Z, Yates J 3rd, Herron B, Otto M, Zan H, Fu H, Casali P
Nature structural & molecular biology 2010 Sep;17(9):1124-35
Nature structural & molecular biology 2010 Sep;17(9):1124-35
E2A-positive gastric MALT lymphoma has weaker plasmacytoid infiltrates and stronger expression of the memory B-cell-associated miR-223: possible correlation with stage and treatment response.
Liu TY, Chen SU, Kuo SH, Cheng AL, Lin CW
Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2010 Nov;23(11):1507-17
Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2010 Nov;23(11):1507-17
Increased targeting of donor switch region and IgE in Sgamma1-deficient B cells.
Misaghi S, Garris CS, Sun Y, Nguyen A, Zhang J, Sebrell A, Senger K, Yan D, Lorenzo MN, Heldens S, Lee WP, Xu M, Wu J, DeForge L, Sai T, Dixit VM, Zarrin AA
Journal of immunology (Baltimore, Md. : 1950) 2010 Jul 1;185(1):166-73
Journal of immunology (Baltimore, Md. : 1950) 2010 Jul 1;185(1):166-73
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
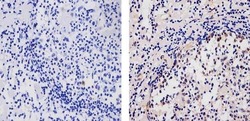
- Immunohistochemistry analysis of AID showing staining in the cytoplasm and nucleus of paraffin-embedded human lymph node tissue (right) compared to a negative control without primary antibody (left). To expose target proteins, antigen retrieval was performed using 10mM sodium citrate (pH 6.0), microwaved for 8-15 min. Following antigen retrieval, tissues were blocked in 3% H2O2-methanol for 15 min at room temperature, washed with ddH2O and PBS, and then probed with a AID Mouse Monoclonal Antibody (Product # 39-2500) diluted in 3% BSA-PBS at a dilution of 1:20 for 1 hour at 37ºC in a humidified chamber. Tissues were washed extensively in PBST and detection was performed using an HRP-conjugated secondary antibody followed by colorimetric detection using a DAB kit. Tissues were counterstained with hematoxylin and dehydrated with ethanol and xylene to prep for mounting.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- Figure 5 14-3-3 adaptors colocalize with AID and RPA in the nucleus of switching B cells. ( a ) Formation of 14-3-3 nuclear foci that colocalized with AID nuclear foci in human 4B6 B cells. ( b ) Formation of 14-3-3 nuclear foci that colocalized with RPA nuclear foci in human 4B6 B cells. ( c ) Formation of 14-3-3 nuclear foci that colocalized with AID nuclear foci in mouse primary B cells stimulated with LPS plus mIL-4 (to induce CSR to IgG1) for 48 hours. ( d ) Formation of 14-3-3 nuclear foci that colocalized with RPA nuclear foci in mouse primary B cells stimulated with LPS plus mIL-4 for 48 hours. Scale bars: 5 mum. Data are representative of those from three independent experiments.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
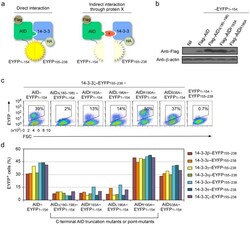
- Figure 1 14-3-3 adaptors interact with AID through the AID C-terminus. ( a ) Schematics of the principle of the BiFC assays to analyze interaction of 14-3-3 (HA-14-3-3-EYFP155-238) and AID (Flag-AID-EYFP1-154). ( b ) Immunoblotting using specific mAbs to identify Flag and beta-actin in HeLa cell expressing nil (pcDNA3 vector), Flag-AID, Flag-AIDDelta(180-198), Flag-AIDF193A, Flag-AIDR190A (fused to EYFP1-154). ( c ) BiFC assays of the interaction between 14-3-3zeta (fused to EYFP155-238) and AID, and AIDR190A and AIDS38A, but not AIDDelta(180-198), AIDF193A or AIDL196A (fused to EYFP1-154) in HeLa cells (at 24 hours), as analyzed by flow cytometry. ( d ) Quantification of the interaction between each of the seven 14-3-3 isoforms (beta, epsilon, gamma, eta, sigma, tau, zeta; fused to EYFP155-238) and AID, AIDDelta(180-198), AIDF193A, AIDL196A, AIDR190A or AIDS38A (fused to EYFP1-154), in HeLa cells (at 48 hours) depicted as percentage of EYFP + , as analyzed by flow cytometry. Data are representative of those from three independent experiments.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
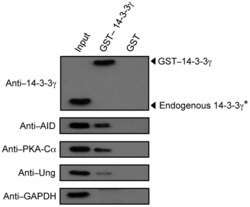
- Figure 3 14-3-3gamma interacts with AID, PKA-Calpha and Ung expressed in B cells undergoing CSR. Immunoblotting using specific mAbs/Abs was performed to identify 14-3-3gamma, AID, PKA-Calpha and Ung in proteins pulled-down with GST-14-3-3gamma or GST alone from whole lysates of mouse B cells stimulated with LPS plus mIL-4 for 48 hours. *Endogenous 14-3-3gamma was detected when a higher amount of GST-14-3-3gamma-precipitated proteins were analyzed, but no signal was detected for AID, PKA or Ung from GST-precipitated proteins. Data are representative of those from three independent experiments.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Figure 7 Inhibition of CSR and disruption of 14-3-3 recruitment to S regions by Vpr. ( a ) Proportions of IgG1 + cells in mouse primary B cells transduced with pTAC-GFP-Vpr or pTAC-GFP retrovirus (which mediated expression of GFP-Vpr or GFP, respectively) and stimulated with LPS plus mIL-4 for 72 hours (left; to induce CSR to IgG1), and quantification of class-switched surface IgG1 + B cells expressing GFP (right; mean and s.e.m of data from three independent experiments), as analyzed by flow cytometry. ( b ) Levels of Aicda , germline Imu-Cmu and Igamma1-Cgamma1, circle Igamma1-Cmu and mature post-recombination Imu-Cgamma1 transcripts in mouse primary B cells expressing GFP-Vpr or GFP and stimulated with LPS plus mIL-4 for 48 hours. Data were normalized to the level of Gapdh transcripts and depicted as the ratio of expression of transcripts in B cells expressing GFP-Vpr to that in B cells expressing GFP (mean and s.e.m of data from three independent experiments). **, P
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
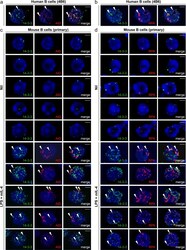
- Figure 10 AID colocalized with Vpr in germinal center B cells from HIV-1 + patients. ( a ) Immunohistochemistry analysis of Vpr in germinal centers in the tonsil from an HIV-1 - subject (left) or in lymph nodes from an HIV-1 + patient (right). Original magnification 100x. ( b ) Immunohistochemistry analysis of Vpr and CD20 (top) or Vpr and AID (bottom) in the tonsil from an HIV-1 - subject (left) or in lymph nodes from an HIV-1 + patient (right). Original magnification 100X. ( c ) Immunofluorescence staining and confocal microscopy analysis of Vpr in human 4B6 B cells cocultured with nil (left), CEM.NK R -CCR5 T cells (middle) or CEM.NK R -CCR5 T cells infected with HIV-192US657 (right). Human 4B6 B cells and CEM.NK R -CCR5 T cells were stained by FITC-conjugated anti-mu Ab (top) and anti-CD4 mAb (bottom), respectively. DAPI (blue) was used to visualize the nucleus. Scale bars: 5 mum. Data are representative of those from three independent experiments.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
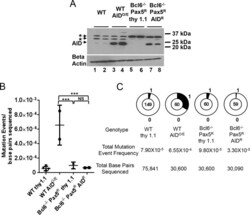
- Fig 1 Pax5 expression does not restore SHM/GCV in Bcl6-deficient DT40 cells. A) Western blot analysis for AID expression in the subclones analyzed for mutations in panels B and C with a beta actin loading control shown below. *, background bands. B) Scatter plot of IgL V region SHM/GCV event frequencies from individual subclones as indicated. Horizontal bars indicate the mean frequency of mutations for each genotype. Z test used to assess significant differences among mutation frequencies. ***, p
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
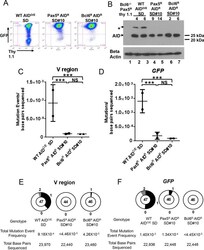
- Fig 3 Super DIVAC does not potentiate mutation in the absence of Bcl6. A) Representative flow cytometry plots of GFP and Thy 1.1 levels after 28 days of culture of the indicated cell lines. B) Western blot analysis for AID expression in the sublcones analyzed for mutation in panels C, D, E, and F, with beta actin loading control shown below. * background band. Each WT subclone contains a unique Super DIVAC GFP2 reporter integration event while all Pax5 R AID R SD#10 and Bcl6 R AID R SD#10 subclones share an identical reporter integration site. C, D) Scatter plots showing the SHM/GCV event frequencies observed at the IgL V region (C) or GFP (D) for 2 independent subclones as indicated after 76 days of culture, presented as in Fig 1B . Horizontal bars indicate the mean frequency of mutations for each genotype. E, F) Pie charts showing the distribution of all SHM/GCV events observed at the IgL V region (E) or GFP (F) compiled from the 2 independent subclones in panels C, D after 76 days of culture, presented as in Fig 1C . Z test was used to assess significance differences among mutation frequencies. ***, p
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
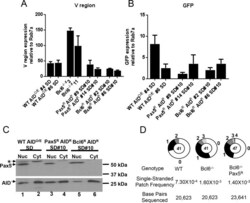
- Fig 4 Transcription, AID subcellular localization, and single stranded DNA in Bcl6-deficient DT40 cells. A, B) RT-PCR analysis of A) IgL V region or B) GFP RNA levels in the sequenced subclones from Fig 3C-3F . Data are presented as the average of the signal obtained from two independent RNA preparations for each subclone after normalization to the signal obtained for Rab7a. Error bars represent the SEM. C) Western blot analysis for AID (lower panel) and Pax5 (upper panel) protein in cytoplasmic and nuclear fractions for the indicated cell lines. All fractions were quantitated and equivalent amounts of protein were loaded. *, unknown band. D) Pie charts showing the distribution of single stranded patches detected in the IgL V region after bisulfite treatment and sequence analysis for one subclone of each genotype. Bisulfite converts dCs on exposed single-stranded DNA to dUs and, after PCR amplification and sequencing, C to T transitions and G to A transitions reveal single-stranded dCs on the top and bottom strands, respectively. To be counted as a single-stranded DNA patch, at least two successive transition mutations were required.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
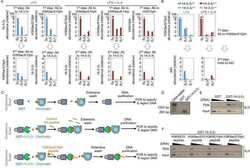
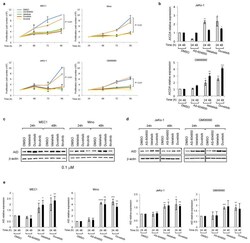
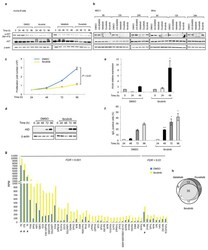
- Extended Data Fig 6 PI3Kdelta blockade increases AID expression in human B cell lines a, Viable cells were counted at the indicated time points by Trypan Blue exclusion in MEC1, Mino, JeKo-1 and GM06990 cell lines treated with the indicated inhibitors (1 muM). Data are expressed as mean +- s.d. (n=3). P values calculated by two-tailed Student's t -test. b , Histograms showing the AICDA mRNA relative expression in JeKo-1 and GM06990 cell lines treated with 1 muM inhibitors. Data are expressed as mean +- s.d. (n = 3 technical replicates, n = 3 biological replicates). * P < 0.05, ** P
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Extended Data Fig 8 Translocations to AID off-targets are increased by idelalisib and duvelisib treatment in MEC1 and JeKo-1 cell line Distribution of translocation junctions in the IgH locus ( a ) and in the IRF4 AID off-target gene ( b ) in MEC1 cells. AID knock-out (KO) MEC1 cells were generated by CRISPR/Cas9 mediated deletion. Numbers of translocation junctions in focal clusters are indicated in bold. c , Translocation junction frequency in AID on-target and AID off-target sites in JeKo-1 B cell line treated with DMSO, idelalisib or duvelisib (1 muM). Data are from pooled HTGTS libraries of similar size ( Supplementary Table 1 and 4 ) from 3 independent experiments. Statistical analysis in Methods. * FDR
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- Extended Data Fig 9 Ibrutinib increases AID expression and the frequency of translocations to AID on- and off-target sites in mouse activated B cells a , AKT phosphorylation was detected by Western Blot in mouse activated B cells treated with DMSO, idelalisib, duvelisib or ibrutinib (1 muM) for the indicated time points (n = 2 biological replicates). For gel source data, see Supplementary Figure 1 . b , MEC1 and Mino human lymphoma cells were treated with the indicated inhibitors (1 muM) and AKT phosphorylation was evaluated by Western Blot (n = 3 biological replicates). c, Viable cells were counted at the indicated time points by Trypan Blue exclusion in activated B cells treated with DMSO or ibrutinib (1 muM). Data are expressed as mean +- s.d. (n=3). P values calculated by two-tailed Student's t -test. d, Western blot for AID protein expression in activated B cells treated with 1 muM ibrutinib. The DMSO panel from Fig. 1a is shown for comparison (n=3 biological replicates). e, Aicda mRNA levels analyzed by qRT-PCR in activated B cells treated with DMSO or ibrutinib (1 muM). Data are expressed as mean +- s.d. (n = 3 technical replicates, n = 3 biological replicates). * P < 0.05, two-tailed Student's t -test. f, IgG 1 CSR in activated B cells analyzed by flow cytometry. Data are expressed as mean +- s.d. (n = 3 biological replicates). * P < 0.05, *** P
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
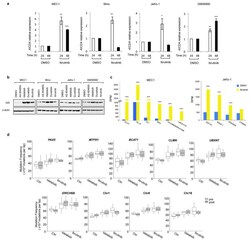
- Extended Data Fig 10 Ibrutinib increases AID expression and the frequency of translocations to AID on- and off-target sites in human B cells a, AICDA mRNA levels analyzed by qRT-PCR in MEC1, Mino, JeKo-1 and GM06990 cell lines after treatment with DMSO or ibrutinib. Data are expressed as mean +- s.d. (n = 3 technical replicates, n = 3 biological replicates). * P < 0.05, ** P
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
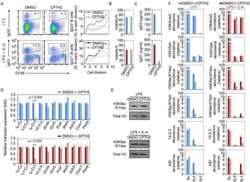
- Figure 4 PI3Kdelta inhibitors increase AID expression and genomic instability in human B cells a,b AICDA mRNA relative expression in the human MEC1 or Mino B cell lines treated with the indicated inhibitors (1 muM). Data are mean +- s.d. (n=3 technical replicates, n = 3 biological replicates). ** P
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
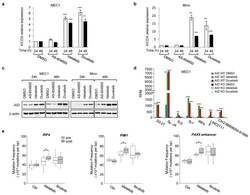
- Extended Data Figure 1 Phosphatidylinositol 3-Kinase (PI3K)delta blockade increases AID expression and CSR in activated mouse B cells a, Schematic representation of the in vivo experiment in WT mice immunized with sheep red blood cells (SRBCs) and treated with the indicated inhibitors (n = 6 biological replicates). b , Representative dot plots of IgG 1 switched GL7 + /B220 + germinal center (GC) B cells from mice treated with vehicle or the PI3Kdelta inhibitors. c, Mean IgG 1 CSR analyzed by flow cytometry in control and idelalisib or duvelisib treated mice. Data are expressed as mean +- s.d. (n=3). ** P
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Extended Data Figure 2 Frequency of AID mediated translocation junctions in activated mouse B cells is increased by PI3Kdelta blockade a , Frequency of chromosomal translocations between the c-myc and the Igh and Igk loci, represented as reads per million (RPM) in WT and AID -/- activated B cells treated with the indicated inhibitors. Significance is calculated as false discovery rate ( FDR ) by comparing AS-604850, idelalisib or duvelisib to DMSO treated B cells as indicated in the Methods. ** FDR
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
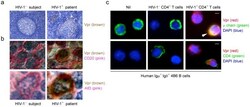
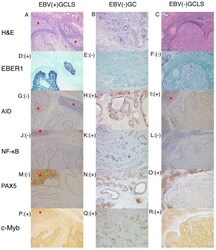
- Figure 1. Representative histology, ISH and immunostaining images of EBV(+) GCLS, EBV(-) GC without LS and EBV(-) GCLS. H&E staining of (A) EBV(+) GCLS, (B) EBV(-) GC without LS and (C) EBV(-) GCLS. EBER1 ISH of (D) EBV(+) GCLS, (E) EBV(-) GC without LS and (F) EBV(-) GCLS. AID immunostaining of (G) EBV(+) GCLS, (H) EBV(-) GC without LS and (I) EBV(-) GCLS. NF-kappaB immunostaining of (J) EBV(+) GCLS, (K) EBV(-) GC without LS and (L) EBV(-) GCLS. PAX5 immunostaining of (M) EBV(+) GCLS, (N) EBV(-) GC without LS and (O) EBV(-) GCLS. c-Myb immunostaining of (P) EBV(+) GCLS, (Q) EBV(-) GC without LS and (R) EBV(-) GCLS. Magnification, x100. (G, I) Red stars indicate AID expression as the positive internal control observed in the lymphoid cells of the germinal center. ISH, in situ hybridization; EBV, Epstein-Barr virus; GCLS, GC with LS; GC, gastric carcinoma; LS, lymphoid stroma; H&E, hematoxylin and eosin; EBER1, EBV-encoded small RNA 1; AID, activation-induced cytidine deaminase; NF-kappaB, nuclear factor kappaB; PAX5, paired box 5; c-Myb, c-Myb proto-oncogene.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
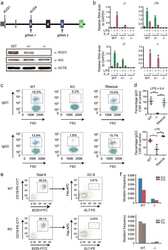
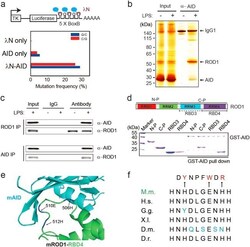
- Fig. 1 ROD1 physically interacts with AID via an ultraconserved loop region. a Diagram of the lamdaN/BoxB tethering assay and the mutation frequency observed in HEK293 cells. The C/G mutations to all C/G bases in BoxB region were calculated from 20 sequenced clones. b Silver staining of AID immunoprecipitates from lysates of either LPS-activated or naive splenic B cells. c ROD1 and AID interact with each other in LPS-activated B cells. The reciprocal co-IP was probed with anti-AID and anti-ROD1 antibodies. d Direct interaction between AID and ROD1 truncated proteins by GST pull-down assay. RRM RNA recognition motif, N-P N-terminal protein, C-P C-terminal protein, RBD3 RNA-binding domain 3, RBD4 RNA-binding domain 4. e The 3D interacting surface of AID (cyan) and ROD1 (green) modeled by PRISM. The key interacting amino acids are labeled in blue and indicated by arrowheads. f The residue composition and conservation of the loop region in ROD1. Amino acids from 504 to 513 were aligned across the animal kingdom. The mutated amino acids at each position are listed and marked by arrowheads. D.r. zebrafish, D.m. fly, X.I. frog, G.g. chicken, H.s. human, M.m. mouse
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
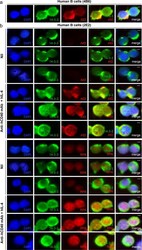
- Figure 4 14-3-3 adaptors codistribute with AID and RPA in the nucleus of switching B cells. ( a ) Codistribution of 14-3-3 and AID in human 4B6 B cells (spontaneous CSR). ( b ) Codistribution of 14-3-3 with AID or RPA in human 2E2 B cells stimulated with agonistic anti-hCD40 mAb plus hIL-4 (to induce CSR to IgG1) for 48 hours. Scale bars: 5 mum. Data are representative of those from three independent experiments.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
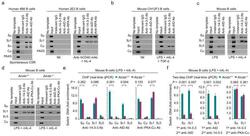
- Figure 2 14-3-3 adaptors bind to the S regions involved in ongoing CSR. ( a ) ChIP assays involving chromatin from human IgM + IgD + 4B6 B cells and Ab to 14-3-3 or AID, and specifying Smu, Sgamma1, Salpha (Salpha1/Salpha2), Cmu and PAX5 DNA (left); or chromatin from human 2E2 B cells stimulated by nil or mAb to hCD40 plus hIL-4, and specifying Smu, Sgamma1, Cmu and PAX5 DNA precipitated by Ab to 14-3-3, AID or RPA (right). ( b,c ) ChIP assays involving chromatin from mouse IgM + CH12F3 B cells stimulated by nil or LPS, mIL-4 plus TGF-beta and specifying Smu, Sgamma1, Salpha and Cmu DNA ( b ), or chromatin from mouse primary B cells (B220 + ) stimulated by LPS or LPS plus mIL-4 and specifying Smu, Sgamma1, Sgamma3 and Cmu DNA ( c ), precipitated by Ab to 14-3-3 or AID. ( d,e ) ChIP assays involving chromatin from Aicda +/+ and Aicda -/- mouse B cells stimulated by LPS plus mIL-4 and Ab to 14-3-3, AID or PKA-Calpha, and specifying ( d ) and quantifying Smu, Sgamma1, Sgamma3 and Cmu DNA ( e , data expressed as ""fold enrichment"" over DNA precipitated by irrelevant IgG, mean and s.d. of triplicate samples). ( f ) Two-step ChIP assays involving chromatin from Aicda +/+ and Aicda -/- mouse B cells stimulated by LPS plus mIL-4 and sequential precipitation by Abs to 14-3-3 and AID (left), or Abs to AID and 14-3-3 (middle), or Abs to 14-3-3 and PKA-Calpha (right). Quantified Smu and Sgamma1 DNA are expressed as fold enrichment over DNA precipitated by irrelevant IgG used in the seco
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
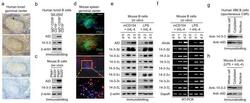
- Figure 3 14-3-3 proteins are upregulated in germinal center B cells and in B cells undergoing CSR in vitro . ( a ) Immunohistochemistry of serial human tonsil sections stained with Ab to 14-3-3, AID or BCL6 (brown signals) or no Ab (""Nil"", showing blue signals from the hematoxylin counterstaining). Scale bar: 50 mum. ( b ) Expression of AID and 14-3-3 in sorted tonsil IgD + CD38 - CD19 + naive and IgD - CD38 + CD19 + germinal center B cells. ( c ) Expression of AID and 14-3-3 in sorted spleen PNA lo B220 + non-germinal center B cells and PNA hi B220 + germinal center B cells from NP 16 -CGG-immunized C57BL/6 mice. ( d ) Expression of 14-3-3 and AID in germinal centers (green, as stained by FITC-conjugated PNA) in serial spleen sections of C57BL/6 mice immunized with NP 16 -CGG for 7 d (one representative of four experiments). The magnified yellow-boxed area shows co-expression of 14-3-3 and AID in germinal center B cells (3 representative cells are indicated by arrowheads). Scale bar: 50 mum. ( e,f ) Immunoblotting ( e ) and RT-PCR ( f ) analysis of AID and the seven 14-3-3 isoforms in freshly isolated mouse primary B cells (d 0) or B cells after stimulation by mCD154 or LPS plus mIL-4. ( g and h ) Expression of 14-3-3 and AID in the whole cell, cytoplasm and nucleus of human 4B6 B cells ( g ) and mouse primary B cells stimulated by LPS plus mIL-4 for 60 h ( h ).
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
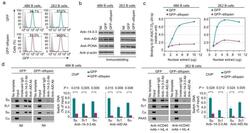
- Figure 4 Blocking 14-3-3 by difopein inhibits 14-3-3 binding to 5'-AGCT-3' repeats and hampers the binding of 14-3-3 and AID to S regions. ( a ) Flow cytometry analysis of the proportion (GFP + ) of human 4B6 and 2E2 B cells expressing nil (gray shade), or GFP or GFP-difopein by lentivirus transduction. ( b ) Expression of 14-3-3 (all isoforms), AID and PCNA in 4B6 B cells and in mAb to hCD40 plus hIL-4-stimulated 2E2 B cells expressing nil, or GFP or GFP-difopein. ( c ) EMSA analysis of the binding to the [5'-AGCT-3'] 3 -24 bp oligonucleotide by two-fold increasing amounts of nuclear extracts from 4B6 B cells and mAb to hCD40 plus hIL-4-stimulated 2E2 B cells expressing GFP or GFP-difopein. Depicted are the quantified and normalized signals of complex A and A' consisting of 14-3-3 and [5'-AGCT-3'] 3 -24 bp. ( d ) ChIP assays involving crosslinked chromatin from 4B6 B cells expressing GFP or GFP-difopein and Ab to 14-3-3 or AID, and specifying Smu, Sgamma1, Salpha (Salpha1/Salpha2) and Cmu region DNA (left panels); and ChIP assays involving chromatin from 2E2 B cells expressing GFP or GFP-difopein after stimulation by mAb to hCD40 plus hIL-4, and specifying Smu, Sgamma1, Cmu and PAX5 DNA after precipitation by Ab to 14-3-3 or AID (right panels). DNA signals were quantified using ImageJ. Depicted is the percentage of input S region DNA precipitated by Ab to 14-3-3 or AID (mean and s.e.m. of three independent experiments).
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
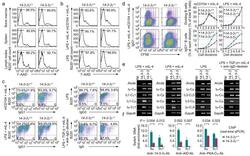
- Figure 6 14-3-3gamma deficiency impaired CSR in mouse primary B cells. ( a,b ) Flow cytometry analysis of viability of 14-3-3gamma + / + and 14-3-3gamma -/- B cells (B220 + ) from bone marrow, spleen and lymph nodes ( a ), and spleen B cells after stimulation by mCD154 or LPS plus mIL-4, or LPS alone ( b ). ( c ) Flow cytometry analysis of CSR in 14-3-3gamma + / + and 14-3-3gamma -/- B cells (one representative of three experiments). The mean and s.e.m. of CSR to each Ig isotype in 14-3-3gamma + / + vs 14-3-3gamma -/- B cells were as follows: mCD154/mIL-4-induced CSR to IgG1, 22.5+-2.7% vs 10.2+-0.5%; LPS/mIL-4-induced CSR to IgG1, 39.8+-4.1% vs 15.2+-4.3%; LPS-induced CSR to IgG3, 7.4+-0.6% vs 3.4%+-0.7%; and LPS, TGF-beta, mIL-4 plus anti-IgD-dextran-induced CSR to IgA, 9.5+-2.7% vs 4.1+-0.8%. ( d ) Flow cytometry analysis of CFSE intensity and surface IgG1 expression in 14-3-3gamma + / + and 14-3-3gamma -/- B cells labeled with CFSE and stimulated by mCD154 or LPS plus mIL-4 for 4 d (left panels). Proportion of B cells completing each cell division (proliferation) and the proportion of surface IgG1 + B cells at each cell division are depicted (right panels). ( e ) RT-PCR analysis of germline I H -C H , circle I H -Cmu and post-recombination Imu-C H transcripts in 14-3-3gamma + / + and 14-3-3gamma -/- B cells stimulated by LPS plus mIL-4, LPS, or LPS, TGF-beta, mIL-4 plus anti-IgD-dextran. Nil: PCR reactions without DNA templates. ( f ) ChIP assays involving chromatin from
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
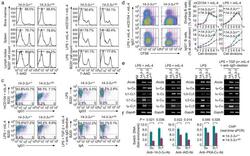
- Figure 7 14-3-3sigma Er DN mutant impaired CSR in mouse primary B cells. ( a,b ) Flow cytometry analysis of viability of 14-3-3sigma + / + and 14-3-3sigma + /Er B cells (B220 + ) from bone marrow, spleen and lymph nodes ( a ), and spleen B cells after stimulation by mCD154 or LPS plus mIL-4, or LPS alone ( b ). ( c ) Flow cytometry analysis of CSR in 14-3-3sigma + / + and 14-3-3sigma + /Er B cells (one representative of three experiments). The mean and s.e.m. of CSR to each Ig isotype in 14-3-3sigma + / + vs 14-3-3sigma + /Er B cells were as follows: mCD154/mIL-4-induced CSR to IgG1, 10.3+-1.9% vs 6.0+-1.8%; LPS/mIL-4-induced CSR to IgG1, 17.5+-2.1% vs 7.2+-1.3%; LPS-induced CSR to IgG3, 5.4+-0.2% vs 2.4%+-0.5%; and LPS, TGF-beta, mIL-4 plus anti-IgD-dextran-induced CSR to IgA, 11.5+-2.7% vs 5.8+-1.9%. ( d ) Flow cytometry analysis of CFSE intensity and surface IgG1 expression in 14-3-3sigma + / + and 14-3-3sigma + /Er B cells labeled with CFSE and stimulated by mCD154 or LPS plus mIL-4 for 4 d (left panels). Proportion of B cells completing each cell division (proliferation) and the proportion of surface IgG1 + B cells at each cell division are depicted (right panels). ( e ) RT-PCR analysis of germline I H -C H , circle I H -Cmu and post-recombination Imu-C H transcripts in 14-3-3sigma + / + and 14-3-3sigma + /Er B cells stimulated by LPS plus mIL-4, LPS, or LPS, TGF-beta, mIL-4 plus anti-IgD-dextran. ( f ) ChIP assays involving chromatin from 14-3-3sigma + / + and 14-3-3sig
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
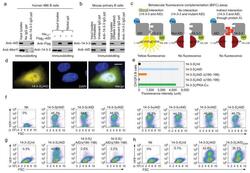
- Figure 8 14-3-3 adaptors interact directly with AID. ( a ) Immunoprecipitation of human 4B6 B cell lysates by rabbit Ab to 14-3-3 followed by immunoblotting with mouse mAb to AID or Mre11 (left); and pull-down of 14-3-3 with AID double-tagged with His 10 -Flag 2 by Ni-NTA-conjugated agarose beads (right). ( b ) Immunoprecipitation of lysates of mouse primary B cells (stimulated by LPS plus mIL-4 for 48 h) by rabbit Ab to 14-3-3 followed by immunoblotting with mouse mAb to 14-3-3 or AID (left). Pre-treatment of lysates with DNase I yielded comparable results (right). ( c ) Schematics of the principle of the BiFC assay. ( d ) BiFC assays of the direct interaction of 14-3-3zeta and AID in HeLa cell nucleus, as revealed by fluorescence microscopy (also presented in Supplementary Fig. 7c ). ( e ) BiFC assays of the direct interaction of 14-3-3zeta with PKA-Calpha and AID, but not AIDDelta(190-198) or AIDDelta(180-198) in mouse CH12F3 B cells stimulated by LPS, mIL-4 plus TGF-beta, as quantified by spectrofluorometry. ( f ) BiFC assays of the direct interaction of each of the seven 14-3-3 isoforms (fused to sEYFP 155-238 ) with AID (fused to sEYFP 1-154 ) in HeLa cells. Nil: co-expression of HA-sEYFP 155-238 and Flag-sEYFP 1-154 , as analyzed by flow cytometry. ( g ) BiFC assays of the direct interaction of 14-3-3zeta with AID, but not AIDDelta(190-198) or AIDDelta(180-198) in HeLa cells (one representative of four experiments). ( h ) BiFC assays of the direct interaction of 14-3-3
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
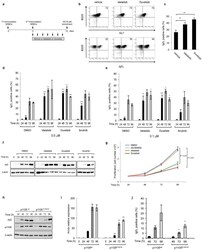
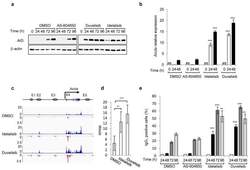
- Figure 1 Phosphatidylinositol 3-Kinase (PI3K)delta blockade increases AID expression and CSR in activated mouse B cells a , Western blot for AID protein from B cells treated with the indicated inhibitors (1 muM) (n = 3 biological replicates). For gel source data, see Supplementary Figure 1 . b , Aicda mRNA levels were analyzed by qRT-PCR. Data are expressed as mean +- s.d. (n = 3 biological replicates).. ** P
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
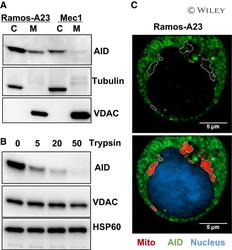
- 4 Fig. Physical separation of AID from mtDNA. (A) Western blot analysis of cytosol and mitochondrial AID proteins from Ramos-A23 and Mec1 cells, showing that a fraction of AID is associated with mitochondria. The purity of proteins was assessed by cytosol-specific Tubulin and mitochondrial outer membrane protein VDAC. (B) Mitochondrial-associated AID is attached to the mitochondrial outer membrane. Purified Ramos-A23 mitochondria were treated with different concentrations of trypsin (0-50 mug*mL -1 ) for 30 min on ice before immunoblot analysis. The integrated mitochondrial outer membrane protein VDAC and mitochondrial matrix protein HSP60 were used as control. All immunoblots were representative of three independent experiments. (C) Confocal microscopy analysis of AID-mitochondria colocalization. Ramos-A23 cells were stained with AID antibody (green) and mitochondrial dye MitoTracker Red (red). Nucleus was counterstained with DAPI (blue). Mitochondrial contours were outlined at the bottom panel and overlaid onto the top panel. The marked mitochondrial areas were essentially free of AID staining. Scale bars: 5 um. C, cytosol protein; M, mitochondrial protein.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot ELISA
ELISA Immunohistochemistry
Immunohistochemistry