Antibody data
- Antibody Data
- Antigen structure
- References [7]
- Comments [0]
- Validations
- Immunocytochemistry [1]
- Immunohistochemistry [1]
- Chromatin Immunoprecipitation [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA000264 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA000264, RRID:AB_611466
- Product name
- Anti-ETV6
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human ETV6, Gene description: ets variant 6, Alternative Gene Names: TEL, Validated applications: ICC, IHC, WB, ChIP, Uniprot ID: P41212, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.1 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references ETV6 Deficiency Unlocks ERG-Dependent Microsatellite Enhancers to Drive Aberrant Gene Activation in B-Lymphoblastic Leukemia.
High ETV6 Levels Support Aggressive B Lymphoma Cell Survival and Predict Poor Outcome in Diffuse Large B-Cell Lymphoma Patients
Regulome analysis in B-acute lymphoblastic leukemia exposes Core Binding Factor addiction as a therapeutic vulnerability
Chromatin-informed inference of transcriptional programs in gynecologic and basal breast cancers
Genome-wide repression of eRNA and target gene loci by the ETV6-RUNX1 fusion in acute leukemia.
Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia
From Gene Expression Analysis to Tissue Microarrays
Kodgule R, Goldman JW, Monovich AC, Saari T, Aguilar AR, Hall CN, Rajesh N, Gupta J, Chu SA, Ye L, Gurumurthy A, Iyer A, Brown NA, Chiang MY, Cieslik MP, Ryan RJH
Blood cancer discovery 2023 Jan 6;4(1):34-53
Blood cancer discovery 2023 Jan 6;4(1):34-53
High ETV6 Levels Support Aggressive B Lymphoma Cell Survival and Predict Poor Outcome in Diffuse Large B-Cell Lymphoma Patients
Marino D, Pizzi M, Kotova I, Schmidt R, Schröder C, Guzzardo V, Talli I, Peroni E, Finotto S, Scapinello G, Dei Tos A, Piazza F, Trentin L, Zagonel V, Piovan E
Cancers 2022;14(2):338
Cancers 2022;14(2):338
Regulome analysis in B-acute lymphoblastic leukemia exposes Core Binding Factor addiction as a therapeutic vulnerability
Wray J, Deltcheva E, Boiers C, Richardson S, Chhetri J, Brown J, Gagrica S, Guo Y, Illendula A, Martens J, Stunnenberg H, Bushweller J, Nimmo R, Enver T
Nature Communications 2022;13(1)
Nature Communications 2022;13(1)
Chromatin-informed inference of transcriptional programs in gynecologic and basal breast cancers
Osmanbeyoglu H, Shimizu F, Rynne-Vidal A, Alonso-Curbelo D, Chen H, Wen H, Yeung T, Jelinic P, Razavi P, Lowe S, Mok S, Chiosis G, Levine D, Leslie C
Nature Communications 2019;10(1)
Nature Communications 2019;10(1)
Genome-wide repression of eRNA and target gene loci by the ETV6-RUNX1 fusion in acute leukemia.
Teppo S, Laukkanen S, Liuksiala T, Nordlund J, Oittinen M, Teittinen K, Grönroos T, St-Onge P, Sinnett D, Syvänen AC, Nykter M, Viiri K, Heinäniemi M, Lohi O
Genome research 2016 Nov;26(11):1468-1477
Genome research 2016 Nov;26(11):1468-1477
Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia
Noetzli L, Lo R, Lee-Sherick A, Callaghan M, Noris P, Savoia A, Rajpurkar M, Jones K, Gowan K, Balduini C, Pecci A, Gnan C, De Rocco D, Doubek M, Li L, Lu L, Leung R, Landolt-Marticorena C, Hunger S, Heller P, Gutierrez-Hartmann A, Xiayuan L, Pluthero F, Rowley J, Weyrich A, Kahr W, Porter C, Di Paola J
Nature Genetics 2015;47(5):535-538
Nature Genetics 2015;47(5):535-538
From Gene Expression Analysis to Tissue Microarrays
Ek S, Andréasson U, Hober S, Kampf C, Pontén F, Uhlén M, Merz H, Borrebaeck C
Molecular & Cellular Proteomics 2006;5(6):1072-1081
Molecular & Cellular Proteomics 2006;5(6):1072-1081
No comments: Submit comment
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
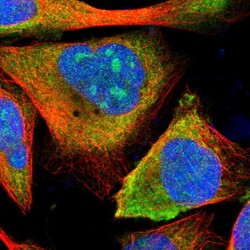
- Experimental details
- Immunofluorescent staining of human cell line U-2 OS shows localization to nucleoli & cytosol.
- Sample type
- Human
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Enhanced method
- Orthogonal validation
- Main image
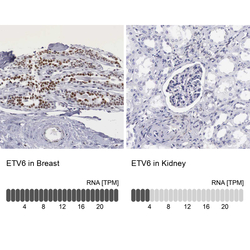
- Experimental details
- Immunohistochemistry analysis in human breast and kidney tissues using HPA000264 antibody. Corresponding ETV6 RNA-seq data are presented for the same tissues.
- Sample type
- Human
- Protocol
- Protocol
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
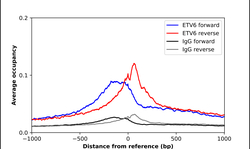
- Experimental details
- ChIP-Exo-Seq composite graph for Anti-ETV6 (HPA000264, Lot 000045525) tested in K562 cells. Strand-specific reads (blue: forward, red: reverse) and IgG controls (black: forward, grey: reverse) are plotted against the distance from a composite set of reference binding sites. The antibody exhibits robust target enrichment compared to a non-specific IgG control and precisely reveals its structural organization around the binding site. Data generated by Prof. B. F. Pugh´s Lab at Cornell University.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry