HPA023535
antibody from Atlas Antibodies
Targeting: SON
BASS1, C21orf50, DBP-5, FLJ21099, FLJ33914, KIAA1019, NREBP
Antibody data
- Antibody Data
- Antigen structure
- References [19]
- Comments [0]
- Validations
- Immunocytochemistry [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA023535 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA023535, RRID:AB_1857362
- Product name
- Anti-SON
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human SON, Gene description: SON DNA binding protein, Alternative Gene Names: BASS1, C21orf50, DBP-5, FLJ21099, FLJ33914, KIAA1019, NREBP, Validated applications: ICC, IHC, Uniprot ID: P18583, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.1 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references Autonomous transposons tune their sequences to ensure somatic suppression
Nuclear RNA-related processes modulate the assembly of cytoplasmic RNA granules
HIV-1 usurps mixed-charge domain-dependent CPSF6 phase separation for higher-order capsid binding, nuclear entry and viral DNA integration
Inter-chromosomal contacts demarcate genome topology along a spatial gradient
Genome-wide analysis of the interplay between chromatin-associated RNA and 3D genome organization in human cells.
Spatial organization of transcribed eukaryotic genes
SANS (USH1G) regulates pre-mRNA splicing by mediating the intra-nuclear transfer of tri-snRNP complexes
The p53-induced RNA-binding protein ZMAT3 is a splicing regulator that inhibits the splicing of oncogenic CD44 variants in colorectal carcinoma
SON and SRRM2 are essential for nuclear speckle formation
Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition
Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler.
Components of a mammalian protein disaggregation/refolding machine are targeted to nuclear speckles following thermal stress in differentiated human neuronal cells
Differential Targeting of Hsp70 Heat Shock Proteins HSPA6 and HSPA1A with Components of a Protein Disaggregation/Refolding Machine in Differentiated Human Neuronal Cells following Thermal Stress
Casein kinase 1 is recruited to nuclear speckles by FAM83H and SON
Dynamics of the association of heat shock protein HSPA6 (Hsp70B’) and HSPA1A (Hsp70–1) with stress-sensitive cytoplasmic and nuclear structures in differentiated human neuronal cells
The conserved ubiquitin-like protein Hub1 plays a critical role in splicing in human cells
Antibody-based Protein Profiling of the Human Chromosome 21
Clustering phenotype populations by genome‐wide RNAi and multiparametric imaging
Ilık İ, Glažar P, Tse K, Brändl B, Meierhofer D, Müller F, Smith Z, Aktaş T
Nature 2024;626(8001):1116-1124
Nature 2024;626(8001):1116-1124
Nuclear RNA-related processes modulate the assembly of cytoplasmic RNA granules
Angel M, Fleshler E, Atrash M, Kinor N, Benichou J, Shav-Tal Y
Nucleic Acids Research 2024;52(9):5356-5375
Nucleic Acids Research 2024;52(9):5356-5375
HIV-1 usurps mixed-charge domain-dependent CPSF6 phase separation for higher-order capsid binding, nuclear entry and viral DNA integration
Jang S, Bedwell G, Singh S, Yu H, Arnarson B, Singh P, Radhakrishnan R, Douglas A, Ingram Z, Freniere C, Akkermans O, Sarafianos S, Ambrose Z, Xiong Y, Anekal P, Montero Llopis P, KewalRamani V, Francis A, Engelman A
Nucleic Acids Research 2024;52(18):11060-11082
Nucleic Acids Research 2024;52(18):11060-11082
Inter-chromosomal contacts demarcate genome topology along a spatial gradient
Mokhtaridoost M, Chalmers J, Soleimanpoor M, McMurray B, Lato D, Nguyen S, Musienko V, Nash J, Espeso-Gil S, Ahmed S, Delfosse K, Browning J, Barutcu A, Wilson M, Liehr T, Shlien A, Aref S, Joyce E, Weise A, Maass P
Nature Communications 2024;15(1)
Nature Communications 2024;15(1)
Genome-wide analysis of the interplay between chromatin-associated RNA and 3D genome organization in human cells.
Calandrelli R, Wen X, Charles Richard JL, Luo Z, Nguyen TC, Chen CJ, Qi Z, Xue S, Chen W, Yan Z, Wu W, Zaleta-Rivera K, Hu R, Yu M, Wang Y, Li W, Ma J, Ren B, Zhong S
Nature communications 2023 Oct 16;14(1):6519
Nature communications 2023 Oct 16;14(1):6519
Kumar A, Schrader A, Boroojeny A, Asadian M, Lee J, Song Y, Zhao S, Han H, Sinha S
2023
2023
Spatial organization of transcribed eukaryotic genes
Leidescher S, Ribisel J, Ullrich S, Feodorova Y, Hildebrand E, Galitsyna A, Bultmann S, Link S, Thanisch K, Mulholland C, Dekker J, Leonhardt H, Mirny L, Solovei I
Nature Cell Biology 2022;24(3):327-339
Nature Cell Biology 2022;24(3):327-339
SANS (USH1G) regulates pre-mRNA splicing by mediating the intra-nuclear transfer of tri-snRNP complexes
Yildirim A, Mozaffari-Jovin S, Wallisch A, Schäfer J, Ludwig S, Urlaub H, Lührmann R, Wolfrum U
Nucleic Acids Research 2021;49(10):5845-5866
Nucleic Acids Research 2021;49(10):5845-5866
The p53-induced RNA-binding protein ZMAT3 is a splicing regulator that inhibits the splicing of oncogenic CD44 variants in colorectal carcinoma
Muys B, Anastasakis D, Claypool D, Pongor L, Li X, Grammatikakis I, Liu M, Wang X, Prasanth K, Aladjem M, Lal A, Hafner M
Genes & Development 2021;35(1-2):102-116
Genes & Development 2021;35(1-2):102-116
SON and SRRM2 are essential for nuclear speckle formation
Ilik İ, Malszycki M, Lübke A, Schade C, Meierhofer D, Aktaş T
eLife 2020;9
eLife 2020;9
Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition
Kim J, Han K, Khanna N, Ha T, Belmont A
Journal of Cell Science 2019;132(8)
Journal of Cell Science 2019;132(8)
Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler.
Chen Y, Zhang Y, Wang Y, Zhang L, Brinkman EK, Adam SA, Goldman R, van Steensel B, Ma J, Belmont AS
The Journal of cell biology 2018 Nov 5;217(11):4025-4048
The Journal of cell biology 2018 Nov 5;217(11):4025-4048
Components of a mammalian protein disaggregation/refolding machine are targeted to nuclear speckles following thermal stress in differentiated human neuronal cells
Deane C, Brown I
Cell Stress and Chaperones 2017;22(2):191-200
Cell Stress and Chaperones 2017;22(2):191-200
Differential Targeting of Hsp70 Heat Shock Proteins HSPA6 and HSPA1A with Components of a Protein Disaggregation/Refolding Machine in Differentiated Human Neuronal Cells following Thermal Stress
Deane C, Brown I
Frontiers in Neuroscience 2017;11
Frontiers in Neuroscience 2017;11
Casein kinase 1 is recruited to nuclear speckles by FAM83H and SON
Kuga T, Kume H, Adachi J, Kawasaki N, Shimizu M, Hoshino I, Matsubara H, Saito Y, Nakayama Y, Tomonaga T
Scientific Reports 2016;6(1)
Scientific Reports 2016;6(1)
Dynamics of the association of heat shock protein HSPA6 (Hsp70B’) and HSPA1A (Hsp70–1) with stress-sensitive cytoplasmic and nuclear structures in differentiated human neuronal cells
Shorbagi S, Brown I
Cell Stress and Chaperones 2016;21(6):993-1003
Cell Stress and Chaperones 2016;21(6):993-1003
The conserved ubiquitin-like protein Hub1 plays a critical role in splicing in human cells
Ammon T, Mishra S, Kowalska K, Popowicz G, Holak T, Jentsch S
Journal of Molecular Cell Biology 2014;6(4):312-323
Journal of Molecular Cell Biology 2014;6(4):312-323
Antibody-based Protein Profiling of the Human Chromosome 21
Uhlén M, Oksvold P, Älgenäs C, Hamsten C, Fagerberg L, Klevebring D, Lundberg E, Odeberg J, Pontén F, Kondo T, Sivertsson Å
Molecular & Cellular Proteomics 2012;11(3):M111.013458
Molecular & Cellular Proteomics 2012;11(3):M111.013458
Clustering phenotype populations by genome‐wide RNAi and multiparametric imaging
Fuchs F, Pau G, Kranz D, Sklyar O, Budjan C, Steinbrink S, Horn T, Pedal A, Huber W, Boutros M
Molecular Systems Biology 2010;6(1)
Molecular Systems Biology 2010;6(1)
No comments: Submit comment
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
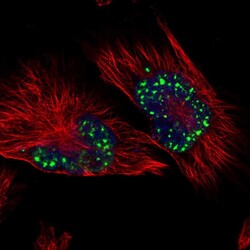
- Experimental details
- Immunofluorescent staining of human cell line U-251 MG shows localization to nuclear speckles.
- Sample type
- Human
 Explore
Explore Validate
Validate Learn
Learn Immunocytochemistry
Immunocytochemistry Immunohistochemistry
Immunohistochemistry