Antibody data
- Antibody Data
- Antigen structure
- References [1]
- Comments [0]
- Validations
- Other assay [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-97639 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- CTCFL Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant full-length protein
- Reactivity
- Human, Mouse
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μg
- Concentration
- 3 mg/mL
- Storage
- -20°C or -80°C if preferred
Submitted references CTCF Expression and Dynamic Motif Accessibility Modulates Epithelial-Mesenchymal Gene Expression.
Johnson KS, Hussein S, Chakraborty P, Muruganantham A, Mikhail S, Gonzalez G, Song S, Jolly MK, Toneff MJ, Benton ML, Lin YC, Taube JH
Cancers 2022 Jan 1;14(1)
Cancers 2022 Jan 1;14(1)
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
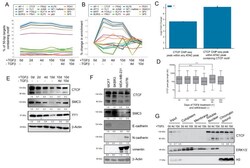
- Figure 4 EMT-driven TFBS accessibility and CTCF expression dynamics. ( A ) Motif enrichment by percentage of 50-bp targets containing motif of the top 20 differentially enriched motifs. ( B ) Motif enrichment percentage change, compared to 0 d, of top 20 differentially enriched motifs. ( C ) The overlap between ATAC-seq peaks (in total, or with CTCF motifs) and CTCF ChIP-seq peaks from Fritz et al. [] was compared to a random distribution model. The observed amount of base pair overlap between the all ATAC-seq peaks and ChIP-seq peaks is 2,758,444 bp, whereas the expected amount of overlap based on our distribution is 110,000.50 bp (s.d. = 4894.19 bp). For the ATAC peaks with a CTCF binding motif, the enrichment is greater (f.c. = 143, p = 0.001). We observe 1,728,041 bp, but expect 12,083.21 bp with a randomly distributed set of genomic regions (s.d. = 1607.12 bp) One thousand permutations were used to generate a p -value, which was corrected using Bonferroni method. * p
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot ELISA
ELISA Other assay
Other assay