Antibody data
- Antibody Data
- Antigen structure
- References [26]
- Comments [0]
- Validations
- Immunohistochemistry [1]
- Chromatin Immunoprecipitation [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA003259 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA003259, RRID:AB_1079381
- Product name
- Anti-MITF
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human MITF, Gene description: microphthalmia-associated transcription factor, Alternative Gene Names: bHLHe32, MI, WS2, WS2A, Validated applications: ChIP, IHC, WB, Uniprot ID: O75030, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.1 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references
DNA damage remodels the MITF interactome to increase melanoma genomic instability
Comparative Analysis of Olive-Derived Phenolic Compounds’ Pro-Melanogenesis Effects on B16F10 Cells and Epidermal Human Melanocytes
A chronic signaling TGFb zebrafish reporter identifies immune response in melanoma
Salvage of ribose from uridine or RNA supports glycolysis in nutrient-limited conditions
Lycopene inhibits endothelial‐to‐mesenchymal transition of choroidal vascular endothelial cells in laser‐induced mouse choroidal neovascularization
A cellular hierarchy in melanoma uncouples growth and metastasis
MITF activity is regulated by a direct interaction with RAF proteins in melanoma cells
The Critical Role Played by Mitochondrial MITF Serine 73 Phosphorylation in Immunologically Activated Mast Cells
Human cutaneous interfollicular melanocytes differentiate temporarily under genotoxic stress.
Activation of the integrated stress response confers vulnerability to mitoribosome-targeting antibiotics in melanoma
TOP1 modulation during melanoma progression and in adaptative resistance to BRAF and MEK inhibitors
BRN2 is a non-canonical melanoma tumor-suppressor
MITF reprograms the extracellular matrix and focal adhesion in melanoma
Classification and Grading of Melanocytic Lesions in a Mouse Model of NRAS-driven Melanomagenesis.
MITF is a driver oncogene and potential therapeutic target in kidney angiomyolipoma tumors through transcriptional regulation of CYR61
PRL3-DDX21 Transcriptional Control of Endolysosomal Genes Restricts Melanocyte Stem Cell Differentiation.
Molecular phenotyping and image-guided surgical treatment of melanoma using spectrally distinct ultrasmall core-shell silica nanoparticles
MITF controls the TCA cycle to modulate the melanoma hypoxia response
The MITF-SOX10 regulated long non-coding RNA DIRC3 is a melanoma tumour suppressor
A direct link between MITF, innate immunity, and hair graying
Lysosomal adaptation: How cells respond to lysosomotropic compounds
BMP-induced reprograming of the retina into RPE requires WNT signalling in the developing chick optic cup
MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment.
Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition
Chang J, Campbell-Hanson K, Vanneste M, Bartschat N, Nagel R, Arnadottir A, Nhung Vu H, Montgomery C, Yevdash J, Jiang J, Bhinu A, Helverson A, Henry M, Steingrímsson E, Weigel R, Cornell R, Kenny C
2025
2025
DNA damage remodels the MITF interactome to increase melanoma genomic instability
Binet R, Lambert J, Tomkova M, Tischfield S, Baggiolini A, Picaud S, Sarkar S, Louphrasitthiphol P, Dias D, Carreira S, Humphrey T, Fillipakopoulos P, White R, Goding C
Genes & Development 2024;38(1-2):70-94
Genes & Development 2024;38(1-2):70-94
Comparative Analysis of Olive-Derived Phenolic Compounds’ Pro-Melanogenesis Effects on B16F10 Cells and Epidermal Human Melanocytes
Cho J, Bejaoui M, Tominaga K, Isoda H
International Journal of Molecular Sciences 2024;25(8):4479
International Journal of Molecular Sciences 2024;25(8):4479
A chronic signaling TGFb zebrafish reporter identifies immune response in melanoma
Thornock A, Noonan H, Barbano J, Xifaras M, Baron C, Yang S, Koczirka K, McConnell A, Zon L
eLife 2024;13
eLife 2024;13
Salvage of ribose from uridine or RNA supports glycolysis in nutrient-limited conditions
Skinner O, Blanco-Fernández J, Goodman R, Kawakami A, Shen H, Kemény L, Joesch-Cohen L, Rees M, Roth J, Fisher D, Mootha V, Jourdain A
Nature Metabolism 2023;5(5):765-776
Nature Metabolism 2023;5(5):765-776
Lycopene inhibits endothelial‐to‐mesenchymal transition of choroidal vascular endothelial cells in laser‐induced mouse choroidal neovascularization
Li L, Cao X, Huang L, Huang X, Gu J, Yu X, Zhu Y, Zhou Y, Song Y, Zhu M
Journal of Cellular and Molecular Medicine 2023;27(10):1327-1340
Journal of Cellular and Molecular Medicine 2023;27(10):1327-1340
Binet R, Lambert J, Tomkova M, Tischfield S, Baggiolini A, Picaud S, Sarkar S, Louphrasitthiphol P, Dias D, Carreira S, Humphrey T, Fillipakopoulos P, White R, Goding C
2023
2023
A cellular hierarchy in melanoma uncouples growth and metastasis
Karras P, Bordeu I, Pozniak J, Nowosad A, Pazzi C, Van Raemdonck N, Landeloos E, Van Herck Y, Pedri D, Bervoets G, Makhzami S, Khoo J, Pavie B, Lamote J, Marin-Bejar O, Dewaele M, Liang H, Zhang X, Hua Y, Wouters J, Browaeys R, Bergers G, Saeys Y, Bosisio F, van den Oord J, Lambrechts D, Rustgi A, Bechter O, Blanpain C, Simons B, Rambow F, Marine J
Nature 2022;610(7930):190-198
Nature 2022;610(7930):190-198
MITF activity is regulated by a direct interaction with RAF proteins in melanoma cells
Estrada C, Mirabal-Ortega L, Méry L, Dingli F, Besse L, Messaoudi C, Loew D, Pouponnot C, Bertolotto C, Eychène A, Druillennec S
Communications Biology 2022;5(1)
Communications Biology 2022;5(1)
The Critical Role Played by Mitochondrial MITF Serine 73 Phosphorylation in Immunologically Activated Mast Cells
Paruchuru L, Govindaraj S, Razin E
Cells 2022;11(3):589
Cells 2022;11(3):589
Human cutaneous interfollicular melanocytes differentiate temporarily under genotoxic stress.
Fessé P, Nyman J, Hermansson I, Book ML, Ahlgren J, Turesson I
iScience 2022 Oct 21;25(10):105238
iScience 2022 Oct 21;25(10):105238
Activation of the integrated stress response confers vulnerability to mitoribosome-targeting antibiotics in melanoma
Vendramin R, Katopodi V, Cinque S, Konnova A, Knezevic Z, Adnane S, Verheyden Y, Karras P, Demesmaeker E, Bosisio F, Kucera L, Rozman J, Gladwyn-Ng I, Rizzotto L, Dassi E, Millevoi S, Bechter O, Marine J, Leucci E
Journal of Experimental Medicine 2021;218(9)
Journal of Experimental Medicine 2021;218(9)
TOP1 modulation during melanoma progression and in adaptative resistance to BRAF and MEK inhibitors
Oliveira É, Chauhan J, Silva J, Carvalho L, Dias D, Carvalho D, Watanabe L, Rebecca V, Mills G, Lu Y, da Silva A, Consolaro M, Herlyn M, Possik P, Goding C, Maria-Engler S
Pharmacological Research 2021;173
Pharmacological Research 2021;173
BRN2 is a non-canonical melanoma tumor-suppressor
Hamm M, Sohier P, Petit V, Raymond J, Delmas V, Le Coz M, Gesbert F, Kenny C, Aktary Z, Pouteaux M, Rambow F, Sarasin A, Charoenchon N, Bellacosa A, Sanchez-del-Campo L, Mosteo L, Lauss M, Meijer D, Steingrimsson E, Jönsson G, Cornell R, Davidson I, Goding C, Larue L
Nature Communications 2021;12(1)
Nature Communications 2021;12(1)
MITF reprograms the extracellular matrix and focal adhesion in melanoma
Dilshat R, Fock V, Kenny C, Gerritsen I, Lasseur R, Travnickova J, Eichhoff O, Cerny P, Möller K, Sigurbjörnsdóttir S, Kirty K, Einarsdottir B, Cheng P, Levesque M, Cornell R, Patton E, Larue L, de Tayrac M, Magnúsdóttir E, Ögmundsdóttir M, Steingrimsson E
eLife 2021;10
eLife 2021;10
Classification and Grading of Melanocytic Lesions in a Mouse Model of NRAS-driven Melanomagenesis.
Assenmacher CA, Santagostino SF, Oyama MA, Marine JC, Bonvin E, Radaelli E
The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society 2021 Mar;69(3):203-218
The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society 2021 Mar;69(3):203-218
MITF is a driver oncogene and potential therapeutic target in kidney angiomyolipoma tumors through transcriptional regulation of CYR61
Zarei M, Giannikou K, Du H, Liu H, Duarte M, Johnson S, Nassar A, Widlund H, Henske E, Long H, Kwiatkowski D
Oncogene 2020;40(1):112-126
Oncogene 2020;40(1):112-126
PRL3-DDX21 Transcriptional Control of Endolysosomal Genes Restricts Melanocyte Stem Cell Differentiation.
Johansson JA, Marie KL, Lu Y, Brombin A, Santoriello C, Zeng Z, Zich J, Gautier P, von Kriegsheim A, Brunsdon H, Wheeler AP, Dreger M, Houston DR, Dooley CM, Sims AH, Busch-Nentwich EM, Zon LI, Illingworth RS, Patton EE
Developmental cell 2020 Aug 10;54(3):317-332.e9
Developmental cell 2020 Aug 10;54(3):317-332.e9
Molecular phenotyping and image-guided surgical treatment of melanoma using spectrally distinct ultrasmall core-shell silica nanoparticles
Chen F, Madajewski B, Ma K, Karassawa Zanoni D, Stambuk H, Turker M, Monette S, Zhang L, Yoo B, Chen P, Meester R, Jonge S, Montero P, Phillips E, Quinn T, Gönen M, Sequeira S, de Stanchina E, Zanzonico P, Wiesner U, Patel S, Bradbury M
Science Advances 2019;5(12)
Science Advances 2019;5(12)
MITF controls the TCA cycle to modulate the melanoma hypoxia response
Louphrasitthiphol P, Ledaki I, Chauhan J, Falletta P, Siddaway R, Buffa F, Mole D, Soga T, Goding C
Pigment Cell & Melanoma Research 2019;32(6):792-808
Pigment Cell & Melanoma Research 2019;32(6):792-808
The MITF-SOX10 regulated long non-coding RNA DIRC3 is a melanoma tumour suppressor
Widlund H, Coe E, Tan J, Shapiro M, Louphrasitthiphol P, Bassett A, Marques A, Goding C, Vance K
PLOS Genetics 2019;15(12):e1008501
PLOS Genetics 2019;15(12):e1008501
A direct link between MITF, innate immunity, and hair graying
Yamashita Y, Harris M, Fufa T, Palmer J, Joshi S, Larson D, Incao A, Gildea D, Trivedi N, Lee A, Day C, Michael H, Hornyak T, Merlino G, Pavan W
PLOS Biology 2018;16(5):e2003648
PLOS Biology 2018;16(5):e2003648
Lysosomal adaptation: How cells respond to lysosomotropic compounds
Komatsu M, Lu S, Sung T, Lin N, Abraham R, Jessen B
PLOS ONE 2017;12(3):e0173771
PLOS ONE 2017;12(3):e0173771
BMP-induced reprograming of the retina into RPE requires WNT signalling in the developing chick optic cup
Steinfeld J, Steinfeld I, Bausch A, Coronato N, Hampel M, Depner H, Layer P, Vogel-Höpker A
Biology Open 2017
Biology Open 2017
MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment.
Riesenberg S, Groetchen A, Siddaway R, Bald T, Reinhardt J, Smorra D, Kohlmeyer J, Renn M, Phung B, Aymans P, Schmidt T, Hornung V, Davidson I, Goding CR, Jönsson G, Landsberg J, Tüting T, Hölzel M
Nature communications 2015 Nov 4;6:8755
Nature communications 2015 Nov 4;6:8755
Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition
Webster D, Barajas B, Bussat R, Yan K, Neela P, Flockhart R, Kovalski J, Zehnder A, Khavari P
Genome Research 2014;24(5):751-760
Genome Research 2014;24(5):751-760
No comments: Submit comment
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Enhanced method
- Orthogonal validation
- Main image
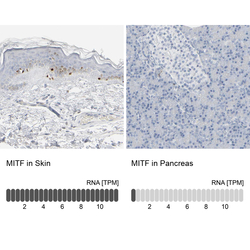
- Experimental details
- Immunohistochemistry analysis in human skin and pancreas tissues using HPA003259 antibody. Corresponding MITF RNA-seq data are presented for the same tissues.
- Sample type
- Human
- Protocol
- Protocol
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
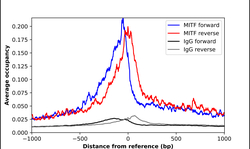
- Experimental details
- ChIP-Exo-Seq composite graph for Anti-MITF (HPA003259, Lot 000014341) tested in K562 cells. Strand-specific reads (blue: forward, red: reverse) and IgG controls (black: forward, grey: reverse) are plotted against the distance from a composite set of reference binding sites. The antibody exhibits robust target enrichment compared to a non-specific IgG control and precisely reveals its structural organization around the binding site. Data generated by Prof. B. F. Pugh´s Lab at Cornell University.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunohistochemistry
Immunohistochemistry