Antibody data
- Antibody Data
- Antigen structure
- References [5]
- Comments [0]
- Validations
- Immunocytochemistry [1]
- Immunohistochemistry [1]
- Chromatin Immunoprecipitation [1]
Submit
Validation data
Reference
Comment
Report error
- Product number
- HPA003916 - Provider product page

- Provider
- Atlas Antibodies
- Proper citation
- Atlas Antibodies Cat#HPA003916, RRID:AB_1857279
- Product name
- Anti-SMARCE1
- Antibody type
- Polyclonal
- Description
- Polyclonal Antibody against Human SMARCE1, Gene description: SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1, Alternative Gene Names: BAF57, Validated applications: ICC, IHC, ChIP, WB, Uniprot ID: Q969G3, Storage: Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Reactivity
- Human, Mouse, Rat
- Host
- Rabbit
- Conjugate
- Unconjugated
- Isotype
- IgG
- Vial size
- 100 µl
- Concentration
- 0.2 mg/ml
- Storage
- Store at +4°C for short term storage. Long time storage is recommended at -20°C.
- Handling
- The antibody solution should be gently mixed before use.
Submitted references Is location more determining than WHO grade for long-term clinical outcome in patients with meningioma in the first two decades of life?
Clear cell meningiomas are defined by a highly distinct DNA methylation profile and mutations in SMARCE1
Loss of SMARCE1 expression is a specific diagnostic marker of clear cell meningioma: a comprehensive immunophenotypical and molecular analysis
Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells
Systematic validation of antibody binding and protein subcellular localization using siRNA and confocal microscopy
Hirschmann D, Nasiri D, Entenmann C, Haberler C, Roetzer T, Dorfer C, Millesi M
Wiener klinische Wochenschrift 2024;137(1-2):21-30
Wiener klinische Wochenschrift 2024;137(1-2):21-30
Clear cell meningiomas are defined by a highly distinct DNA methylation profile and mutations in SMARCE1
Sievers P, Sill M, Blume C, Tauziede-Espariat A, Schrimpf D, Stichel D, Reuss D, Dogan H, Hartmann C, Mawrin C, Hasselblatt M, Stummer W, Schick U, Hench J, Frank S, Ketter R, Schweizer L, Schittenhelm J, Puget S, Brandner S, Jaunmuktane Z, Küsters B, Abdullaev Z, Pekmezci M, Snuderl M, Ratliff M, Herold-Mende C, Unterberg A, Aldape K, Ellison D, Wesseling P, Reifenberger G, Wick W, Perry A, Varlet P, Pfister S, Jones D, von Deimling A, Sahm F
Acta Neuropathologica 2020;141(2):281-290
Acta Neuropathologica 2020;141(2):281-290
Loss of SMARCE1 expression is a specific diagnostic marker of clear cell meningioma: a comprehensive immunophenotypical and molecular analysis
Tauziede‐Espariat A, Parfait B, Besnard A, Lacombe J, Pallud J, Tazi S, Puget S, Lot G, Terris B, Cohen J, Vidaud M, Figarella‐Branger D, Monnien F, Polivka M, Adle‐Biassette H, Varlet P
Brain Pathology 2017;28(4):466-474
Brain Pathology 2017;28(4):466-474
Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells
Stadler C, Rexhepaj E, Singan V, Murphy R, Pepperkok R, Uhlén M, Simpson J, Lundberg E
Nature Methods 2013;10(4):315-323
Nature Methods 2013;10(4):315-323
Systematic validation of antibody binding and protein subcellular localization using siRNA and confocal microscopy
Stadler C, Hjelmare M, Neumann B, Jonasson K, Pepperkok R, Uhlén M, Lundberg E
Journal of Proteomics 2012;75(7):2236-2251
Journal of Proteomics 2012;75(7):2236-2251
No comments: Submit comment
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
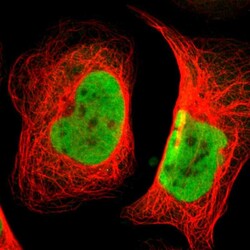
- Experimental details
- Immunofluorescent staining of human cell line U-2 OS shows localization to nucleoplasm.
- Sample type
- Human
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Enhanced method
- Orthogonal validation
- Main image
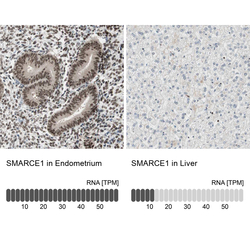
- Experimental details
- Immunohistochemistry analysis in human endometrium and liver tissues using Anti-SMARCE1 antibody. Corresponding SMARCE1 RNA-seq data are presented for the same tissues.
- Sample type
- Human
- Protocol
- Protocol
Supportive validation
- Submitted by
- Atlas Antibodies (provider)
- Main image
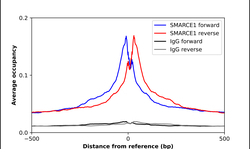
- Experimental details
- ChIP-Exo-Seq composite graph for Anti-SMARCE1 (HPA003916, Lot A107052) tested in K562 cells. Strand-specific reads (blue: forward, red: reverse) and IgG controls (black: forward, grey: reverse) are plotted against the distance from a composite set of reference binding sites. The antibody exhibits robust target enrichment compared to a non-specific IgG control and precisely reveals its structural organization around the binding site. Data generated by Prof. B. F. Pugh´s Lab at Cornell University.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot Immunocytochemistry
Immunocytochemistry