Antibody data
- Antibody Data
- Antigen structure
- References [3]
- Comments [0]
- Validations
- Western blot [3]
- Immunohistochemistry [2]
- Other assay [3]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA5-27395 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- ADAM17 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Recombinant full-length protein
- Description
- Recommended positive controls: HeLa, K562. Predicted reactivity: Mouse (89%), Rat (87%), Dog (89%), Pig (85%), Chicken (80%). Store product as a concentrated solution. Centrifuge briefly prior to opening the vial.
- Reactivity
- Human, Mouse, Rat
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 1.21 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references The E3 ubiquitin-protein ligase Trim31 alleviates non-alcoholic fatty liver disease by targeting Rhbdf2 in mouse hepatocytes.
Cardiac and Renal SARS-CoV-2 Viral Entry Protein Regulation by Androgens and Diet: Implications for Polycystic Ovary Syndrome and COVID-19.
Iron-Overload triggers ADAM-17 mediated inflammation in Severe Alcoholic Hepatitis.
Xu M, Tan J, Dong W, Zou B, Teng X, Zhu L, Ge C, Dai X, Kuang Q, Zhong S, Lai L, Yi C, Tang T, Zhao J, Wang L, Liu J, Wei H, Sun Y, Yang Q, Li Q, Lou D, Hu L, Liu X, Kuang G, Luo J, Xiong M, Feng J, Zhang C, Wang B
Nature communications 2022 Feb 25;13(1):1052
Nature communications 2022 Feb 25;13(1):1052
Cardiac and Renal SARS-CoV-2 Viral Entry Protein Regulation by Androgens and Diet: Implications for Polycystic Ovary Syndrome and COVID-19.
Rezq S, Huffman AM, Basnet J, Yanes Cardozo LL, Romero DG
International journal of molecular sciences 2021 Sep 9;22(18)
International journal of molecular sciences 2021 Sep 9;22(18)
Iron-Overload triggers ADAM-17 mediated inflammation in Severe Alcoholic Hepatitis.
Maras JS, Das S, Sharma S, Sukriti S, Kumar J, Vyas AK, Kumar D, Bhat A, Yadav G, Choudhary MC, Sharma S, Kumar G, Bihari C, Trehanpati N, Maiwall R, Sarin SK
Scientific reports 2018 Jul 6;8(1):10264
Scientific reports 2018 Jul 6;8(1):10264
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
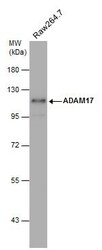
- Experimental details
- Western Blot using ADAM17 Polyclonal Antibody (Product # PA5-27395). Whole cell extract (30 µg) was separated by 7.5% SDS-PAGE, and the membrane was blotted with ADAM17 Polyclonal Antibody (Product # PA5-27395) diluted at 1:500. The HRP-conjugated anti-rabbit IgG antibody was used to detect the primary antibody.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
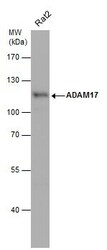
- Experimental details
- Western Blot using ADAM17 Polyclonal Antibody (Product # PA5-27395). Whole cell extract (30 µg) was separated by 7.5% SDS-PAGE, and the membrane was blotted with ADAM17 Polyclonal Antibody (Product # PA5-27395) diluted at 1:500. The HRP-conjugated anti-rabbit IgG antibody was used to detect the primary antibody.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
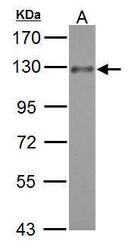
- Experimental details
- ADAM17 Polyclonal Antibody detects ADAM17 protein by western blot analysis. A. 30 µg K562 whole cell lysate/extract .7.5% SDS-PAGE. ADAM17 Polyclonal Antibody (Product # PA5-27395) dilution: 1:500. The HRP-conjugated anti-rabbit IgG antibody was used to detect the primary antibody.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
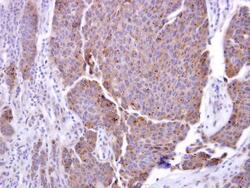
- Experimental details
- ADAM17 Polyclonal Antibody detects ADAM17 protein at cytosol on human breast carcinoma by immunohistochemical analysis. Sample: ADAM17 Polyclonal Antibody (Product # PA5-27395) dilution: 1:250. Antigen Retrieval: EDTA based buffer, pH 8.0, 15 min.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
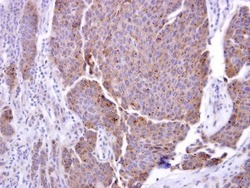
- Experimental details
- ADAM17 Polyclonal Antibody detects ADAM17 protein at cytosol on human breast carcinoma by immunohistochemical analysis. Sample: ADAM17 Polyclonal Antibody (Product # PA5-27395) dilution: 1:250. Antigen Retrieval: EDTA based buffer, pH 8.0, 15 min.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
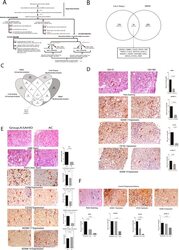
- Experimental details
- Figure 2 Increased CD163 and ADAM17 expression is associated to iron-overload in SAH. ( A ) Workflow and summary of the experiments performed and number of samples used in target determination, and validation experiments including the In-vivo and In-vitro analysis. ( B ) Venn-diagram for unique and shared genes (p < 0.05, fold change >1.5) in the liver and PBMC transcriptome of Group A: SAH-IO.16 shared genes in the liver and PBMC were associated to cellular iron ion homeostasis, cytokine receptor binding and notch signaling. ( C ) Venn-diagram for unique and shared genes identified in the univariate and multivariate projection analysis for the liver and PBMC transcriptome of Group A: SAH-IO documenting CD163, ADAM17 and TMED7 significantly associated to systemic iron overload. ( D ) Immuno-histochemistry (IHC) for iron deposition (Perl's-staining) and expression of ADAM17, CD163, ADAM10 in Group A: SAH-IO (n = 5) vs. Group B: SAH-NIO (n = 5). [For all IHC analysis relative quantization of positively stained cells are expressed as mean number of positive cells/10 high power field (40x)]. ( E ) Perl's staining and expression of CD68, ADAM17, CD163 and ADAM10 in Group A: SAH-IO (n = 5) vs. AC (n = 5). ( F ) Perl's staining, and expression of ADAM17, CD163 and ADAM10 in Thalassemia (n = 3) patients compared to Group A and B.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
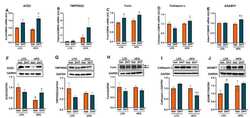
- Experimental details
- Figure 3 Effect of DHT and diet on cardiac SARS-CoV-2 viral entry proteins expression. Animals were treated with dihydrotestosterone (DHT) or vehicle (Veh) and maintained in low (LFD) or high (HFD) fat diet for 90 days. Ace2 ( A ), Tmprss2 ( B ), furin ( C ), cathepsin L ( D ), and ADAM17 ( E ) mRNA was quantified by RT-qPCR and standardized to the geometric mean of four housekeeping genes (HGMK) ( N = 6/group). ACE2 ( F ), TMPRSS2 ( G ), furin ( H ), cathepsin L ( I ), and ADAM17 ( J ) protein was quantified by Western-blot and normalized to GAPDH ( N = 4/group). Total furin ( H ) was quantified as the sum of profurin (dashed arrow) and cleaved soluble furin (solid arrow). Data are expressed as mean +- SEM. Data were analyzed by two-way ANOVA followed by Fisher's LSD test. a p < 0.05 vs. LFD-Veh; b p < 0.05 vs. HFD-Veh; c p < 0.05 vs. LFD-DHT.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
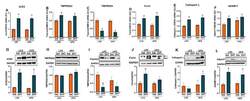
- Experimental details
- Figure 4 Effect of DHT and diet on renal SARS-CoV-2 viral entry proteins expression. Animals were treated with dihydrotestosterone (DHT) or vehicle (Veh) and maintained in low (LFD) or high (HFD) fat diet for 90 days. Ace2 ( A ), Tmprss2 ( B ), Tmprss4 ( C ), furin ( D ), cathepsin L ( E ), and ADAM17 ( F ) mRNA was quantified by RT-qPCR and standardized to the geometric mean of four housekeeping genes (HGMK) ( N = 6-8/group). ACE2 ( G ), TMPRSS2 ( H ), TMPRSS4 ( I ), furin ( J ), cathepsin L ( K ), and ADAM17 ( L ) protein was quantified by Western-blot and normalized to GAPDH ( N = 4/group). Total furin ( J ) was quantified as the sum of profurin (dashed arrow) and cleaved soluble furin (solid arrow). Data are expressed as mean +- SEM. Data were analyzed by two-way ANOVA followed by Fisher's LSD test. a p < 0.05 vs. LFD-Veh; b p < 0.05 vs. LFD-Veh; c p < 0.05 vs. LFD-DHT.
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot