PA1-31402
antibody from Invitrogen Antibodies
Targeting: OGG1
HMMH, HOGG1, MUTM, OGH1
 Western blot
Western blot ELISA
ELISA Immunocytochemistry
Immunocytochemistry Immunoprecipitation
Immunoprecipitation Immunohistochemistry
Immunohistochemistry Flow cytometry
Flow cytometry Other assay
Other assayAntibody data
- Antibody Data
- Antigen structure
- References [4]
- Comments [0]
- Validations
- Immunocytochemistry [2]
- Immunohistochemistry [1]
- Other assay [10]
Submit
Validation data
Reference
Comment
Report error
- Product number
- PA1-31402 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- OGG1 Polyclonal Antibody
- Antibody type
- Polyclonal
- Antigen
- Synthetic peptide
- Description
- Store product as a concentrated solution. Centrifuge briefly prior to opening the vial.
- Reactivity
- Human, Mouse, Rat
- Host
- Rabbit
- Isotype
- IgG
- Vial size
- 100 μL
- Concentration
- 1.0 mg/mL
- Storage
- Store at 4°C short term. For long term storage, store at -20°C, avoiding freeze/thaw cycles.
Submitted references The DNA repair function of CUX1 contributes to radioresistance.
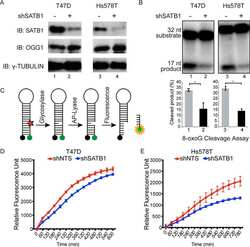
Special AT-rich Sequence-binding Protein 1 (SATB1) Functions as an Accessory Factor in Base Excision Repair.
CUX2 protein functions as an accessory factor in the repair of oxidative DNA damage.
RAS transformation requires CUX1-dependent repair of oxidative DNA damage.
Ramdzan ZM, Ginjala V, Pinder JB, Chung D, Donovan CM, Kaur S, Leduy L, Dellaire G, Ganesan S, Nepveu A
Oncotarget 2017 Mar 21;8(12):19021-19038
Oncotarget 2017 Mar 21;8(12):19021-19038
Special AT-rich Sequence-binding Protein 1 (SATB1) Functions as an Accessory Factor in Base Excision Repair.
Kaur S, Coulombe Y, Ramdzan ZM, Leduy L, Masson JY, Nepveu A
The Journal of biological chemistry 2016 Oct 21;291(43):22769-22780
The Journal of biological chemistry 2016 Oct 21;291(43):22769-22780
CUX2 protein functions as an accessory factor in the repair of oxidative DNA damage.
Pal R, Ramdzan ZM, Kaur S, Duquette PM, Marcotte R, Leduy L, Davoudi S, Lamarche-Vane N, Iulianella A, Nepveu A
The Journal of biological chemistry 2015 Sep 11;290(37):22520-31
The Journal of biological chemistry 2015 Sep 11;290(37):22520-31
RAS transformation requires CUX1-dependent repair of oxidative DNA damage.
Ramdzan ZM, Vadnais C, Pal R, Vandal G, Cadieux C, Leduy L, Davoudi S, Hulea L, Yao L, Karnezis AN, Paquet M, Dankort D, Nepveu A
PLoS biology 2014 Mar;12(3):e1001807
PLoS biology 2014 Mar;12(3):e1001807
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
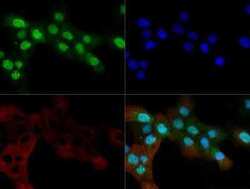
- Experimental details
- Immunocytochemistry-Immunofluorescence analysis of OGG1 in A431 cells. Cells were fixed with 10% formalin, permeabilized with 0.5% Triton-X100 and OGG1 Polyclonal Antibody (Product # PA1-31402) (Green) at a dilution of 1:200 was used as a primary antibody. Red : Tubulin. Blue: DAPI.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
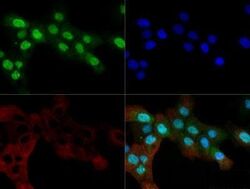
- Experimental details
- Immunocytochemistry-Immunofluorescence analysis of OGG1 in A431 cells. Cells were fixed with 10% formalin, permeabilized with 0.5% Triton-X100 and OGG1 Polyclonal Antibody (Product # PA1-31402) (Green) at a dilution of 1:200 was used as a primary antibody. Red : Tubulin. Blue: DAPI.
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
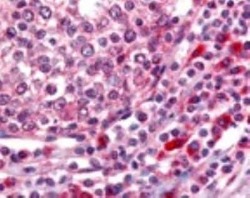
- Experimental details
- Immunohistochemistry (Paraffin) analysis of OGG1 in human tonsillar germinal center and mantle zone tissue using OGG1 Polyclonal Antibody (Product # PA1-31402).
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
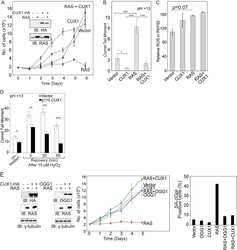
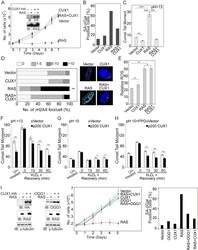
- Figure 3 CUX1 prevents RAS-induced cell senescence. (A) IMR90 cells stably expressing p200 CUX1 or carrying an empty vector were infected with a retrovirus expressing HRAS G12V or an empty vector. Following selection, cells were seeded in triplicate and counted for 7 d. The graph is a representative example of three independent experiments. CUX1-HA and HRAS expression were verified by immunoblotting analyses. (B) The percentage of cells exhibiting SA-betagal activity on day 7 was measured. At least 120 cells were analyzed in each case. (C) On day 6 postselection, IMR90 cells were collected and DNA strand breaks quantified by alkaline (pH>13) single-cell gel electrophoresis. The graph is a representative example of three independent experiments. * Indicates p value
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
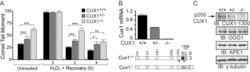
- Figure 5 Genetic inactivation of Cux1 reduces the DNA repair efficiency of MEFs. (A) MEFs from Cux1 +/+ , Cux1 +/- , and Cux1 -/- mice were exposed to 10 um H 2 O 2 for 20 min on ice, allowed to recover at 37degC for the indicated time. DNA damage before and after treatment was measured by comet assay at pH>13 as in Figure 3F , except that the time course was extended since recovery takes longer in MEFs. Each bar represents the average of at least 30 comets. * p
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
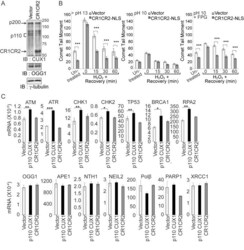
- Figure 6 Acceleration of DNA repair does not require the transcriptional functions of CUX1. (A) DLD-1 cells were stably transfected with a plasmid expressing CUX1 CR1CR2 fused to a nuclear localization signal (NLS), or with empty vector (vector). Expression of recombinant CUX1 protein expression and OGG1 levels were analyzed by immunoblotting. (B) DLD-1 cells were exposed to 10 um H 2 O 2 for 20 min on ice, allowed to recover at 37degC for the indicated time, and then submitted to Single Cell Gel Electrophoresis (comet assay) to quantify DNA damage as described in Figure 3F,G,H . Each bar represents the average of at least 30 comets. * p
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
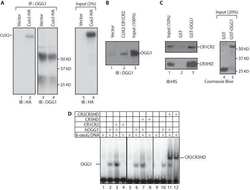
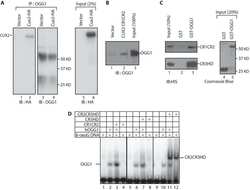
- Figure 7 CUX1 stimulates the DNA glycosylase activity of OGG1. (A) Hs578T cells stably carrying a lentivirus expressing a doxycycline inducible CUX1 shRNA were cultured in the absence (-) or presence of doxycycline for 4 d (+). Expression of CUX1 and OGG1 were analyzed by immunoblotting. (B and C) 8-oxoG cleavage assay was conducted using radiolabeled double-stranded oligonucleotides containing an 8-oxoG (map in Figure S5 ) and 20 ug of whole cell extracts from the indicated cells. (D, Top) Schematic representation of CUX1 proteins used in this study. Shown at the top are the functional domains: Inh, auto-inhibitory domain; CC, coiled-coil; CR1, CR2, and CR3, Cut repeat 1, 2, and 3; HD, cut homeodomain; R1 and R2, repression domains 1 and 2. (D, Bottom) The 8-oxoG cleavage assay was performed using purified human OGG1 and 50 nM of the indicated proteins. ERR FL, full length estrogen-related receptor protein; ERR DBD, DNA binding domain of estrogen-related receptor; CR2CR3HD, CR3HD, and CR1CR2 are CUX1 recombinant proteins described in Figure 6A . (E) Electrophoretic mobility shift assays were performed using oligonucleotides containing an 8-oxoG or an unmodified G and purified OGG1, in the presence or absence of purified CUX1 recombinant proteins, as indicated.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
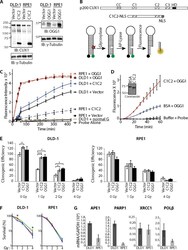
- Figure 5 Cut domains and OGG1 Accelerate Repair of Oxidative DNA Damage and Increase Resistance to Radiation ( A ) DLD-1 (KRAS G13D ) and RPE1 cells were infected with retroviruses expressing either nothing (Vector), C1C2-NLS or OGG1. Nuclear extracts were analyzed by immunoblotting using the indicated antibodies. ( B ) Map showing the evolutionarily conserved domains of CUX1:CC, coiled-coil; C1, C2, C3, CUT domain 1, 2 and 3; HD, homeodomain. The CUT domain 1 and 2 (C1C2) construct includes a nuclear localization signal (NLS). Diagrammatic representation of the 8-oxoG cleavage assay using a fluorophore reporter probe. Red star, 8-oxoguanine; green circle, FAM fluorophore; black circle, dabcyl quencher. ( C ) 8-oxoG cleavage assay was performed using whole cell extracts from DLD-1 and RPE1 cells stably carrying vectors expressing either OGG1, CUT domains 1 and 2 (C1C2) or nothing (vector). Two controls are shown. ""Probe Alone"" is the probe incubated in the absence of cell extract. ""DLD1+normal G"" is a reaction with DLD1 cell extract and a probe that contains a normal guanine residue instead of 8-oxoG; the lack of fluorescence confirms that that the increase in fluorescence intensity in the reactions with the 8-oxoG probe is not caused by a nonspecific nuclease activity. ( D ) 8-oxoG cleavage assay was performed using 50 nM OGG1 and 200 nM of BSA or recombinant CUT domains 1 and 2 (C1C2). A coomassie blue stain of the C1C2 purified protein is shown. ( E ) Cells were expose
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
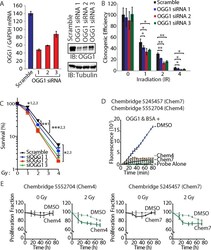
- Figure 7 OGG1 knockdown or inhibition sensitizes cancer cells to radiation ( A ) DLD-1 cells were transfected with three distinct siRNA against OGG1 or with a scramble RNA. OGG1 expression was monitored by RT-qPCR analysis and immunoblotting. ( B ) Cells were submitted to irradiation and their clonogenic efficiency was determined. Cloning efficiency of unexposed cells was set at 100%. Results of triplicate experiments are shown. Error bars represent standard error. *** p < 0.001; ** p < 0.01.; * p < 0.05; Student's t -test. ( C ) Results from clonogenic efficiency data in Figure 7B are represented as line graphs, with all un-irradiated cells set at 100%. Results of triplicate experiments are shown. Error bars represent standard error. *** p < 0.001; ** p < 0.01.; * p < 0.05; Student's t -test comparing each siRNA OGG1 against the scramble RNA control. 1: OGG1 siRNA 1; 2 : OGG1 siRNA 2; 3: OGG1 siRNA 3. ( D ) 8-oxoG cleavage assay was performed using 50 nM OGG1 and BSA in the presence of 10 muM. Chembridge 5552704 and 5245457 compounds or vehicle (DMSO). The two compounds inhibit the reaction. ( E ) DLD-1 colorectal cancer cells were irradiated (0 and 2 Gy) in the presence of 10 muM of Chembridge 5552704 and 5245457 compounds or vehicle (DMSO). Thymidine incorporation was measured for 3 days and is reported relative to the DMSO 0 h value.
 Explore
Explore Validate
Validate Learn
Learn